Intro to Raster Data
Last updated on 2025-11-11 | Edit this page
Overview
Questions
- What is a raster dataset?
- How do I work with and plot raster data in R?
- How can I handle missing or bad data values for a raster?
Objectives
- Describe the fundamental attributes of a raster dataset.
- Explore raster attributes and metadata using R.
- Import rasters into R using the
terrapackage. - Plot a raster file in R using the
ggplot2package. - Describe the difference between single- and multi-band rasters.
Things You’ll Need To Complete This Episode
See the lesson homepage for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
In this episode, we will introduce the fundamental principles, packages and metadata/raster attributes that are needed to work with raster data in R. We will discuss some of the core metadata elements that we need to understand to work with rasters in R, including CRS and resolution. We will also explore missing and bad data values as stored in a raster and how R handles these elements.
We will continue to work with the dplyr and
ggplot2 packages that were introduced in the Introduction to R
for Geospatial Data lesson. We will use two additional packages in
this episode to work with raster data - the terra and
sf packages. Make sure that you have these packages
loaded.
R
library(terra)
library(ggplot2)
library(dplyr)
Introduce the Data
If not already discussed, introduce the datasets that will be used in this lesson. A brief introduction to the datasets can be found on the Geospatial workshop homepage.
For more detailed information about the datasets, check out the Geospatial workshop data page.
View Raster File Attributes
We will be working with a series of GeoTIFF files in this lesson. The
GeoTIFF format contains a set of embedded tags with metadata about the
raster data. We can use the function describe() to get
information about our raster data before we read that data into R. It is
ideal to do this before importing your data.
R
describe("data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_dsmCrop.tif")
OUTPUT
[1] "Driver: GTiff/GeoTIFF"
[2] "Files: data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_dsmCrop.tif"
[3] "Size is 1697, 1367"
[4] "Coordinate System is:"
[5] "PROJCRS[\"WGS 84 / UTM zone 18N\","
[6] " BASEGEOGCRS[\"WGS 84\","
[7] " DATUM[\"World Geodetic System 1984\","
[8] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
[9] " LENGTHUNIT[\"metre\",1]]],"
[10] " PRIMEM[\"Greenwich\",0,"
[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
[12] " ID[\"EPSG\",4326]],"
[13] " CONVERSION[\"UTM zone 18N\","
[14] " METHOD[\"Transverse Mercator\","
[15] " ID[\"EPSG\",9807]],"
[16] " PARAMETER[\"Latitude of natural origin\",0,"
[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[18] " ID[\"EPSG\",8801]],"
[19] " PARAMETER[\"Longitude of natural origin\",-75,"
[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[21] " ID[\"EPSG\",8802]],"
[22] " PARAMETER[\"Scale factor at natural origin\",0.9996,"
[23] " SCALEUNIT[\"unity\",1],"
[24] " ID[\"EPSG\",8805]],"
[25] " PARAMETER[\"False easting\",500000,"
[26] " LENGTHUNIT[\"metre\",1],"
[27] " ID[\"EPSG\",8806]],"
[28] " PARAMETER[\"False northing\",0,"
[29] " LENGTHUNIT[\"metre\",1],"
[30] " ID[\"EPSG\",8807]]],"
[31] " CS[Cartesian,2],"
[32] " AXIS[\"(E)\",east,"
[33] " ORDER[1],"
[34] " LENGTHUNIT[\"metre\",1]],"
[35] " AXIS[\"(N)\",north,"
[36] " ORDER[2],"
[37] " LENGTHUNIT[\"metre\",1]],"
[38] " USAGE["
[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
[40] " AREA[\"Between 78°W and 72°W, northern hemisphere between equator and 84°N, onshore and offshore. Bahamas. Canada - Nunavut; Ontario; Quebec. Colombia. Cuba. Ecuador. Greenland. Haiti. Jamica. Panama. Turks and Caicos Islands. United States (USA). Venezuela.\"],"
[41] " BBOX[0,-78,84,-72]],"
[42] " ID[\"EPSG\",32618]]"
[43] "Data axis to CRS axis mapping: 1,2"
[44] "Origin = (731453.000000000000000,4713838.000000000000000)"
[45] "Pixel Size = (1.000000000000000,-1.000000000000000)"
[46] "Metadata:"
[47] " AREA_OR_POINT=Area"
[48] "Image Structure Metadata:"
[49] " COMPRESSION=LZW"
[50] " INTERLEAVE=BAND"
[51] "Corner Coordinates:"
[52] "Upper Left ( 731453.000, 4713838.000) ( 72d10'52.71\"W, 42d32'32.18\"N)"
[53] "Lower Left ( 731453.000, 4712471.000) ( 72d10'54.71\"W, 42d31'47.92\"N)"
[54] "Upper Right ( 733150.000, 4713838.000) ( 72d 9'38.40\"W, 42d32'30.35\"N)"
[55] "Lower Right ( 733150.000, 4712471.000) ( 72d 9'40.41\"W, 42d31'46.08\"N)"
[56] "Center ( 732301.500, 4713154.500) ( 72d10'16.56\"W, 42d32' 9.13\"N)"
[57] "Band 1 Block=1697x1 Type=Float64, ColorInterp=Gray"
[58] " Min=305.070 Max=416.070 "
[59] " Minimum=305.070, Maximum=416.070, Mean=359.853, StdDev=17.832"
[60] " NoData Value=-9999"
[61] " Metadata:"
[62] " STATISTICS_MAXIMUM=416.06997680664"
[63] " STATISTICS_MEAN=359.85311802914"
[64] " STATISTICS_MINIMUM=305.07000732422"
[65] " STATISTICS_STDDEV=17.83169335933" If you wish to store this information in R, you can do the following:
R
harv_metadata <- capture.output(
describe("data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_dsmCrop.tif")
)
Each line of text that was printed to the console is now stored as an
element of the character vector harv_metadata. We will be
exploring this data throughout this episode. By the end of this episode,
you will be able to explain and understand the output above.
Open a Raster in R
Now that we’ve previewed the metadata for our GeoTIFF, let’s import
this raster dataset into R and explore its metadata more closely. We can
use the rast() function to open a raster in R.
Data Tip - Object names
To improve code readability, file and object names should be used
that make it clear what is in the file. The data for this episode were
collected from Harvard Forest so we’ll use a naming convention of
datatype_HARV.
First we will load our raster file into R and view the data structure.
R
dsm_harv <-
rast("data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_dsmCrop.tif")
dsm_harv
OUTPUT
class : SpatRaster
size : 1367, 1697, 1 (nrow, ncol, nlyr)
resolution : 1, 1 (x, y)
extent : 731453, 733150, 4712471, 4713838 (xmin, xmax, ymin, ymax)
coord. ref. : WGS 84 / UTM zone 18N (EPSG:32618)
source : HARV_dsmCrop.tif
name : HARV_dsmCrop
min value : 305.07
max value : 416.07 The information above includes a report of min and max values, but no other data range statistics. Similar to other R data structures like vectors and data frame columns, descriptive statistics for raster data can be retrieved like
R
summary(dsm_harv)
WARNING
Warning: [summary] used a sampleOUTPUT
HARV_dsmCrop
Min. :305.6
1st Qu.:345.6
Median :359.6
Mean :359.8
3rd Qu.:374.3
Max. :414.7 but note the warning - unless you force R to calculate these
statistics using every cell in the raster, it will take a random sample
of 100,000 cells and calculate from that instead. To force calculation
all the values, you can use the function values:
R
summary(values(dsm_harv))
OUTPUT
HARV_dsmCrop
Min. :305.1
1st Qu.:345.6
Median :359.7
Mean :359.9
3rd Qu.:374.3
Max. :416.1 To visualise this data in R using ggplot2, we need to
convert it to a dataframe. We learned about dataframes in an
earlier lesson. The terra package has an built-in
function for conversion to a plotable dataframe.
R
dsm_harv_df <- as.data.frame(dsm_harv, xy = TRUE)
Now when we view the structure of our data, we will see a standard dataframe format.
R
str(dsm_harv_df)
OUTPUT
'data.frame': 2319799 obs. of 3 variables:
$ x : num 731454 731454 731456 731456 731458 ...
$ y : num 4713838 4713838 4713838 4713838 4713838 ...
$ HARV_dsmCrop: num 409 408 407 407 409 ...We can use ggplot() to plot this data. We will set the
color scale to scale_fill_viridis_c which is a
color-blindness friendly color scale. We will also use the
coord_quickmap() function to use an approximate Mercator
projection for our plots. This approximation is suitable for small areas
that are not too close to the poles. Other coordinate systems are
available in ggplot2 if needed, you can learn about them at their help
page ?coord_map.
R
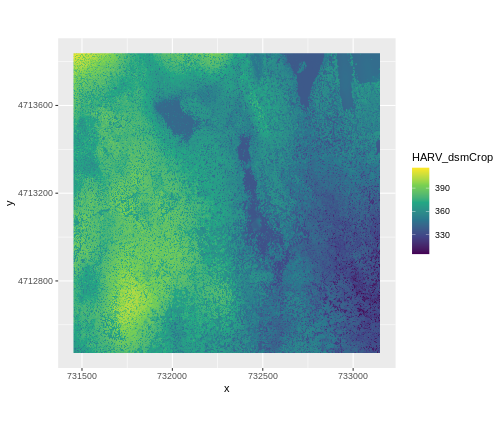
ggplot() +
geom_raster(data = dsm_harv_df , aes(x = x, y = y, fill = HARV_dsmCrop)) +
scale_fill_viridis_c() +
coord_quickmap()
Plotting Tip
More information about the Viridis palette used above at R Viridis package documentation.
Plotting Tip
For faster, simpler plots, you can use the plot function
from the terra package.
See ?plot for more arguments to customize the plot
R
plot(dsm_harv)
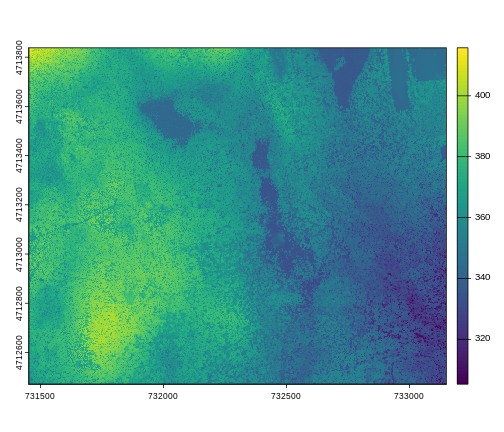
This map shows the elevation of our study site in Harvard Forest. From the legend, we can see that the maximum elevation is ~400, but we can’t tell whether this is 400 feet or 400 meters because the legend doesn’t show us the units.
Unfortunately, nothing in the technical metadata (that which is built into the file format) tells us what the vertical resolution is. For now you will have to trust us that it is meters.
However, we can look at the metadata of our object to see what the horizontal units are: in other words how many meters from the origin are the x and y points. That metadata, is part of the CRS. We introduced the concept of a CRS in an earlier lesson.
Now we will see how features of the CRS appear in our data file and what meanings they have.
View Raster Coordinate Reference System (CRS) in R
We can view the CRS string associated with our R object using
thecrs() function.
R
crs(dsm_harv, proj = TRUE)
OUTPUT
[1] "+proj=utm +zone=18 +datum=WGS84 +units=m +no_defs"Challenge
What units are our horizontal data in?
+units=m tells us that our data is in meters.
Understanding CRS in Proj4 Format
The CRS for our data is given to us by R in proj4
format. Let’s break down the pieces of proj4 string. The
string contains all of the individual CRS elements that R or another GIS
might need. Each element is specified with a + sign,
similar to how a .csv file is delimited or broken up by a
,. After each + we see the CRS element being
defined. For example projection (proj=) and datum
(datum=).
UTM Proj4 String
A projection string (like the one of dsm_harv) specifies
the UTM projection as follows:
+proj=utm +zone=18 +datum=WGS84 +units=m +no_defs +ellps=WGS84 +towgs84=0,0,0
- proj=utm: the projection is UTM, UTM has several zones.
- zone=18: the zone is 18
- datum=WGS84: the datum is WGS84 (the datum refers to the 0,0 reference for the coordinate system used in the projection)
- units=m: the units for the coordinates are in meters
- ellps=WGS84: the ellipsoid (how the earth’s roundness is calculated) for the data is WGS84
Note that the zone is unique to the UTM projection. Not all CRSs will have a zone. Image source: Chrismurf at English Wikipedia, via Wikimedia Commons (CC-BY).
Calculate Raster Min and Max Values
It is useful to know the minimum or maximum values of a raster dataset. In this case, given we are working with elevation data, these values represent the min/max elevation range at our site.
Raster statistics are often calculated and embedded in a GeoTIFF for us. We can view these values:
R
minmax(dsm_harv)
OUTPUT
HARV_dsmCrop
min 305.07
max 416.07R
min(values(dsm_harv))
OUTPUT
[1] 305.07R
max(values(dsm_harv))
OUTPUT
[1] 416.07Data Tip - Set min and max values
If the minimum and maximum values haven’t already been calculated, we
can calculate them using the setMinMax() function.
R
dsm_harv <- setMinMax(dsm_harv)
We can see that the elevation at our site ranges from 305.0700073m to 416.0699768m.
Raster Bands
The Digital Surface Model object (dsm_harv) that we’ve
been working with is a single band raster. This means that there is only
one dataset stored in the raster: surface elevation in meters for one
time period.
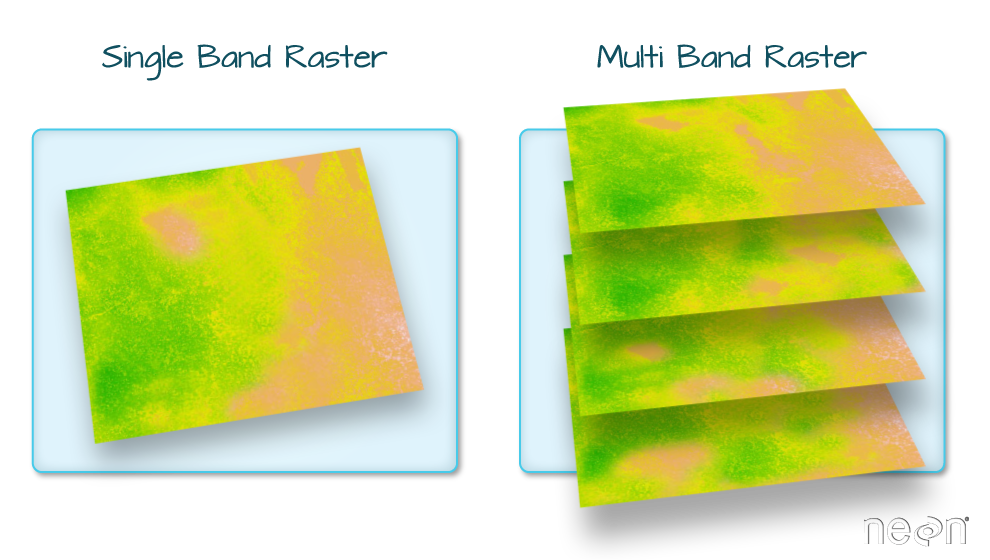
A raster dataset can contain one or more bands. We can use the
rast() function to import one single band from a single or
multi-band raster. We can view the number of bands in a raster using the
nlyr() function.
R
nlyr(dsm_harv)
OUTPUT
[1] 1However, raster data can also be multi-band, meaning that one raster file contains data for more than one variable or time period for each cell. Jump to a later episode in this series for information on working with multi-band rasters: Work with Multi-band Rasters in R.
Dealing with Missing Data
Raster data often has a NoDataValue associated with it.
This is a value assigned to pixels where data is missing or no data were
collected.
By default the shape of a raster is always rectangular. So if we have
a dataset that has a shape that isn’t rectangular, some pixels at the
edge of the raster will have NoDataValues. This often
happens when the data were collected by an airplane which only flew over
some part of a defined region.
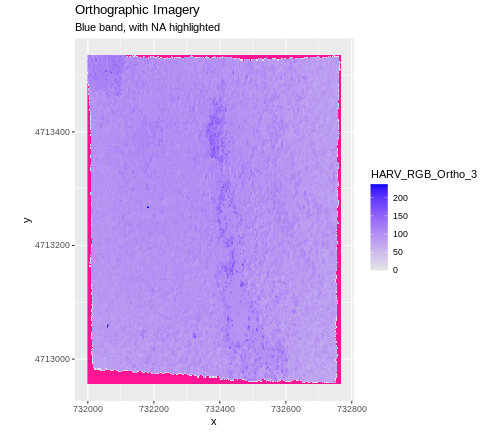
In the image below, the pixels that are black have
NoDataValues. The camera did not collect data in these
areas.
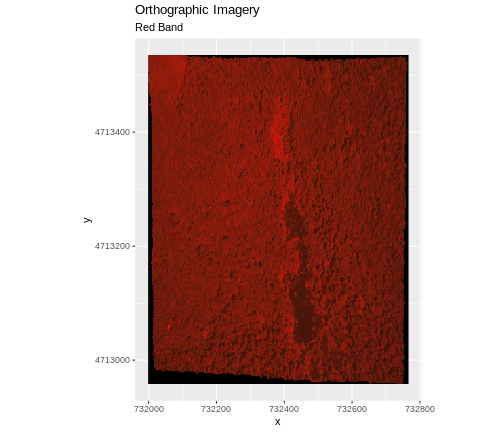
In the next image, the black edges have been assigned
NoDataValue. R doesn’t render pixels that contain a
specified NoDataValue. R assigns missing data with the
NoDataValue as NA.
The difference here shows up as ragged edges on the plot, rather than black spaces where there is no data.

If your raster already has NA values set correctly but
you aren’t sure where they are, you can deliberately plot them in a
particular colour. This can be useful when checking a dataset’s
coverage. For instance, sometimes data can be missing where a sensor
could not ‘see’ its target data, and you may wish to locate that missing
data and fill it in.
To highlight NA values in ggplot, alter the
scale_fill_*() layer to contain a colour instruction for
NA values, like
scale_fill_viridis_c(na.value = 'deeppink')
The value that is conventionally used to take note of missing data
(the NoDataValue value) varies by the raster data type. For
floating-point rasters, the figure -3.4e+38 is a common
default, and for integers, -9999 is common. Some
disciplines have specific conventions that vary from these common
values.
In some cases, other NA values may be more appropriate.
An NA value should be a) outside the range of valid values,
and b) a value that fits the data type in use. For instance, if your
data ranges continuously from -20 to 100, 0 is not an acceptable
NA value! Or, for categories that number 1-15, 0 might be
fine for NA, but using -.000003 will force you to save the
GeoTIFF on disk as a floating point raster, resulting in a bigger
file.
If we are lucky, our GeoTIFF file has a tag that tells us what is the
NoDataValue. If we are less lucky, we can find that
information in the raster’s metadata. If a NoDataValue was
stored in the GeoTIFF tag, when R opens up the raster, it will assign
each instance of the value to NA. Values of NA
will be ignored by R as demonstrated above.
Challenge
Use the output from the describe() and
sources() functions to find out what
NoDataValue is used for our dsm_harv
dataset.
R
describe(sources(dsm_harv))
OUTPUT
[1] "Driver: GTiff/GeoTIFF"
[2] "Files: /home/runner/work/r-raster-vector-geospatial/r-raster-vector-geospatial/site/built/data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_dsmCrop.tif"
[3] "Size is 1697, 1367"
[4] "Coordinate System is:"
[5] "PROJCRS[\"WGS 84 / UTM zone 18N\","
[6] " BASEGEOGCRS[\"WGS 84\","
[7] " DATUM[\"World Geodetic System 1984\","
[8] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
[9] " LENGTHUNIT[\"metre\",1]]],"
[10] " PRIMEM[\"Greenwich\",0,"
[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
[12] " ID[\"EPSG\",4326]],"
[13] " CONVERSION[\"UTM zone 18N\","
[14] " METHOD[\"Transverse Mercator\","
[15] " ID[\"EPSG\",9807]],"
[16] " PARAMETER[\"Latitude of natural origin\",0,"
[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[18] " ID[\"EPSG\",8801]],"
[19] " PARAMETER[\"Longitude of natural origin\",-75,"
[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[21] " ID[\"EPSG\",8802]],"
[22] " PARAMETER[\"Scale factor at natural origin\",0.9996,"
[23] " SCALEUNIT[\"unity\",1],"
[24] " ID[\"EPSG\",8805]],"
[25] " PARAMETER[\"False easting\",500000,"
[26] " LENGTHUNIT[\"metre\",1],"
[27] " ID[\"EPSG\",8806]],"
[28] " PARAMETER[\"False northing\",0,"
[29] " LENGTHUNIT[\"metre\",1],"
[30] " ID[\"EPSG\",8807]]],"
[31] " CS[Cartesian,2],"
[32] " AXIS[\"(E)\",east,"
[33] " ORDER[1],"
[34] " LENGTHUNIT[\"metre\",1]],"
[35] " AXIS[\"(N)\",north,"
[36] " ORDER[2],"
[37] " LENGTHUNIT[\"metre\",1]],"
[38] " USAGE["
[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
[40] " AREA[\"Between 78°W and 72°W, northern hemisphere between equator and 84°N, onshore and offshore. Bahamas. Canada - Nunavut; Ontario; Quebec. Colombia. Cuba. Ecuador. Greenland. Haiti. Jamica. Panama. Turks and Caicos Islands. United States (USA). Venezuela.\"],"
[41] " BBOX[0,-78,84,-72]],"
[42] " ID[\"EPSG\",32618]]"
[43] "Data axis to CRS axis mapping: 1,2"
[44] "Origin = (731453.000000000000000,4713838.000000000000000)"
[45] "Pixel Size = (1.000000000000000,-1.000000000000000)"
[46] "Metadata:"
[47] " AREA_OR_POINT=Area"
[48] "Image Structure Metadata:"
[49] " COMPRESSION=LZW"
[50] " INTERLEAVE=BAND"
[51] "Corner Coordinates:"
[52] "Upper Left ( 731453.000, 4713838.000) ( 72d10'52.71\"W, 42d32'32.18\"N)"
[53] "Lower Left ( 731453.000, 4712471.000) ( 72d10'54.71\"W, 42d31'47.92\"N)"
[54] "Upper Right ( 733150.000, 4713838.000) ( 72d 9'38.40\"W, 42d32'30.35\"N)"
[55] "Lower Right ( 733150.000, 4712471.000) ( 72d 9'40.41\"W, 42d31'46.08\"N)"
[56] "Center ( 732301.500, 4713154.500) ( 72d10'16.56\"W, 42d32' 9.13\"N)"
[57] "Band 1 Block=1697x1 Type=Float64, ColorInterp=Gray"
[58] " Min=305.070 Max=416.070 "
[59] " Minimum=305.070, Maximum=416.070, Mean=359.853, StdDev=17.832"
[60] " NoData Value=-9999"
[61] " Metadata:"
[62] " STATISTICS_MAXIMUM=416.06997680664"
[63] " STATISTICS_MEAN=359.85311802914"
[64] " STATISTICS_MINIMUM=305.07000732422"
[65] " STATISTICS_STDDEV=17.83169335933" NoDataValue are encoded as -9999.
Bad Data Values in Rasters
Bad data values are different from NoDataValues. Bad
data values are values that fall outside of the applicable range of a
dataset.
Examples of Bad Data Values:
- The normalized difference vegetation index (NDVI), which is a measure of greenness, has a valid range of -1 to 1. Any value outside of that range would be considered a “bad” or miscalculated value.
- Reflectance data in an image will often range from 0-1 or 0-10,000 depending upon how the data are scaled. Thus a value greater than 1 or greater than 10,000 is likely caused by an error in either data collection or processing.
Find Bad Data Values
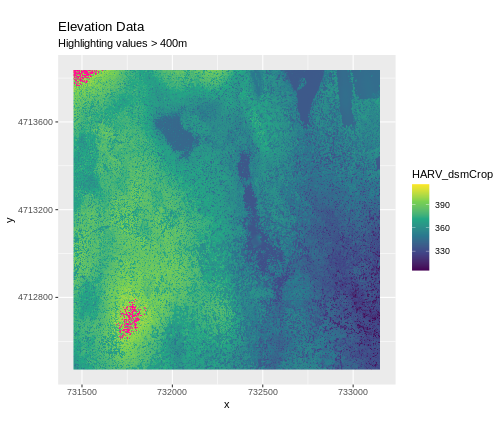
Sometimes a raster’s metadata will tell us the range of expected values for a raster. Values outside of this range are suspect and we need to consider that when we analyze the data. Sometimes, we need to use some common sense and scientific insight as we examine the data - just as we would for field data to identify questionable values.
Plotting data with appropriate highlighting can help reveal patterns in bad values and may suggest a solution. Below, reclassification is used to highlight elevation values over 400m with a contrasting colour.
Create A Histogram of Raster Values
We can explore the distribution of values contained within our raster
using the geom_histogram() function which produces a
histogram. Histograms are often useful in identifying outliers and bad
data values in our raster data.
R
ggplot() +
geom_histogram(data = dsm_harv_df, aes(HARV_dsmCrop))
OUTPUT
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.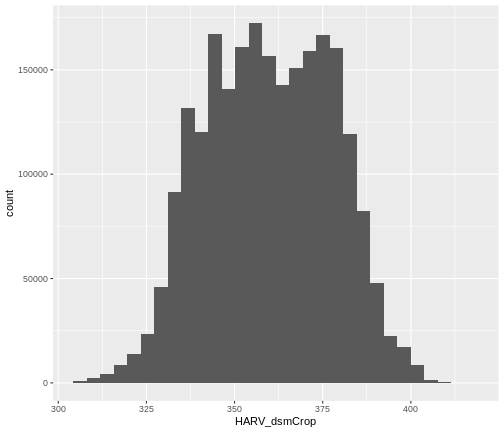
Notice that a warning message is thrown when R creates the histogram.
stat_bin() using bins = 30. Pick better
value with binwidth.
This warning is caused by a default setting in
geom_histogram enforcing that there are 30 bins for the
data. We can define the number of bins we want in the histogram by using
the bins value in the geom_histogram()
function.
R
ggplot() +
geom_histogram(data = dsm_harv_df, aes(HARV_dsmCrop), bins = 40)
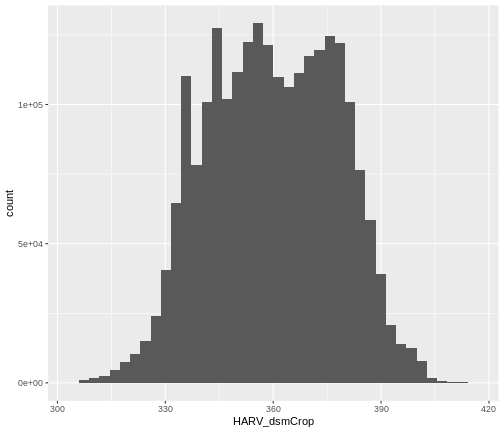
Note that the shape of this histogram looks similar to the previous
one that was created using the default of 30 bins. The distribution of
elevation values for our Digital Surface Model (DSM) looks
reasonable. It is likely there are no bad data values in this particular
raster.
Challenge: Explore Raster Metadata
Use describe() to determine the following about the
NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_DSMhill.tif
file:
- Does this file have the same CRS as
dsm_harv? - What is the
NoDataValue? - What is resolution of the raster data?
- How large would a 5x5 pixel area be on the Earth’s surface?
- Is the file a multi- or single-band raster?
Notice: this file is a hillshade. We will learn about hillshades in the Working with Multi-band Rasters in R episode.
R
describe("data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_DSMhill.tif")
OUTPUT
[1] "Driver: GTiff/GeoTIFF"
[2] "Files: data/NEON-DS-Airborne-Remote-Sensing/HARV/DSM/HARV_DSMhill.tif"
[3] "Size is 1697, 1367"
[4] "Coordinate System is:"
[5] "PROJCRS[\"WGS 84 / UTM zone 18N\","
[6] " BASEGEOGCRS[\"WGS 84\","
[7] " DATUM[\"World Geodetic System 1984\","
[8] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
[9] " LENGTHUNIT[\"metre\",1]]],"
[10] " PRIMEM[\"Greenwich\",0,"
[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
[12] " ID[\"EPSG\",4326]],"
[13] " CONVERSION[\"UTM zone 18N\","
[14] " METHOD[\"Transverse Mercator\","
[15] " ID[\"EPSG\",9807]],"
[16] " PARAMETER[\"Latitude of natural origin\",0,"
[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[18] " ID[\"EPSG\",8801]],"
[19] " PARAMETER[\"Longitude of natural origin\",-75,"
[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[21] " ID[\"EPSG\",8802]],"
[22] " PARAMETER[\"Scale factor at natural origin\",0.9996,"
[23] " SCALEUNIT[\"unity\",1],"
[24] " ID[\"EPSG\",8805]],"
[25] " PARAMETER[\"False easting\",500000,"
[26] " LENGTHUNIT[\"metre\",1],"
[27] " ID[\"EPSG\",8806]],"
[28] " PARAMETER[\"False northing\",0,"
[29] " LENGTHUNIT[\"metre\",1],"
[30] " ID[\"EPSG\",8807]]],"
[31] " CS[Cartesian,2],"
[32] " AXIS[\"(E)\",east,"
[33] " ORDER[1],"
[34] " LENGTHUNIT[\"metre\",1]],"
[35] " AXIS[\"(N)\",north,"
[36] " ORDER[2],"
[37] " LENGTHUNIT[\"metre\",1]],"
[38] " USAGE["
[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
[40] " AREA[\"Between 78°W and 72°W, northern hemisphere between equator and 84°N, onshore and offshore. Bahamas. Canada - Nunavut; Ontario; Quebec. Colombia. Cuba. Ecuador. Greenland. Haiti. Jamica. Panama. Turks and Caicos Islands. United States (USA). Venezuela.\"],"
[41] " BBOX[0,-78,84,-72]],"
[42] " ID[\"EPSG\",32618]]"
[43] "Data axis to CRS axis mapping: 1,2"
[44] "Origin = (731453.000000000000000,4713838.000000000000000)"
[45] "Pixel Size = (1.000000000000000,-1.000000000000000)"
[46] "Metadata:"
[47] " AREA_OR_POINT=Area"
[48] "Image Structure Metadata:"
[49] " COMPRESSION=LZW"
[50] " INTERLEAVE=BAND"
[51] "Corner Coordinates:"
[52] "Upper Left ( 731453.000, 4713838.000) ( 72d10'52.71\"W, 42d32'32.18\"N)"
[53] "Lower Left ( 731453.000, 4712471.000) ( 72d10'54.71\"W, 42d31'47.92\"N)"
[54] "Upper Right ( 733150.000, 4713838.000) ( 72d 9'38.40\"W, 42d32'30.35\"N)"
[55] "Lower Right ( 733150.000, 4712471.000) ( 72d 9'40.41\"W, 42d31'46.08\"N)"
[56] "Center ( 732301.500, 4713154.500) ( 72d10'16.56\"W, 42d32' 9.13\"N)"
[57] "Band 1 Block=1697x1 Type=Float64, ColorInterp=Gray"
[58] " Min=-0.714 Max=1.000 "
[59] " Minimum=-0.714, Maximum=1.000, Mean=0.313, StdDev=0.481"
[60] " NoData Value=-9999"
[61] " Metadata:"
[62] " STATISTICS_MAXIMUM=0.99999973665016"
[63] " STATISTICS_MEAN=0.31255246777216"
[64] " STATISTICS_MINIMUM=-0.71362979358008"
[65] " STATISTICS_STDDEV=0.48129385401108" - If this file has the same CRS as dsm_harv? Yes: UTM Zone 18, WGS84, meters.
- What format
NoDataValuestake? -9999 - The resolution of the raster data? 1x1
- How large a 5x5 pixel area would be? 5mx5m How? We are given resolution of 1x1 and units in meters, therefore resolution of 5x5 means 5x5m.
- Is the file a multi- or single-band raster? Single.
- The GeoTIFF file format includes metadata about the raster data.
- To plot raster data with the
ggplot2package, we need to convert it to a dataframe. - R stores CRS information in the Proj4 format.
- Be careful when dealing with missing or bad data values.
