Learning Objectives
Following this assignment students should be able to:
- understand and use the basic relational operators
- use an
ifstatement to evaluate conditionals- use
ifstatements with functions
Reading
-
Topics
- Conditionals
-
Readings
Lecture Notes
Exercises
Choice Operators (10 pts)
Create the following variables.
w <- 10.2 x <- 1.3 y <- 2.8 z <- 17.5 colors <- c("red", "blue", "green") masses <- c(45.2, 36.1, 27.8, 81.6, 42.4) dna1 <- "attattaggaccaca" dna2 <- "attattaggaacaca"Use them to print whether or not the following statements are
TRUEorFALSE.wis greater than 10"green"is incolorsxis greater thany- Each value in
massesis greater than 40. - 2 *
x+ 0.2 is equal toy dna1is the same asdna2dna1is not the same asdna2wis greater thanx, oryis greater thanzxtimeswis between 13.2 and 13.5 (there is no way to indicated “between” in R so to do this we have to separately check if the number is greater than the minimum value and less than the maximum value, combining these two conditions with&)- Each mass in
massesis between 30 and 50.
Basic If Statements (20 pts)
1. Complete (i.e., copy into your code and them modify) the following
ifstatement so that ifage_classis equal to “sapling” it setsy <- 10.age_class = "sapling" if (){ } y2. Complete the following
ifstatement so that ifage_classis equal to “sapling” it setsy <- 10and ifage_classis equal to “seedling” it setsy <- 5.age_class = "seedling" if (){ } y3. Complete the following
ifstatement so that ifage_classis equal to “sapling” it setsy <- 10and ifage_classis equal to “seedling” it setsy <- 5and ifage_classis something else then it sets the value ofy <- 0.age_class = "adult" if (){ } y4. Convert your
Expected outputs for Basic If Statementsif/else if/elsestatement from (3) into a function that takesage_classas an argument and returnsy. Call this function 5 times, once with each of the following values forage_class: “sapling”, “seedling”, “adult”, “mature”, “established”.If Statements In Functions (20 pts)
1. Write a function named
double_if_smallthat- Takes a number as input
- If the input is less than 26, returns the number multiplied by 2
- If the input is greater than or equal to 26, returns just the original number (not multiplied by two).
Call the function with
10as the input.2. Call the function from (1) with
30as the input.3. Write a function called
predictionthat takes a single argumentx. Ifxis both greater than 0 and less than 15 then returny = 6 + 0.8 * x. Ifxis both greater than 15 and less than 30 then returny = 5 + 0.75 * x. In all other cases returny = NA. Call the function with5as the input.4. Call the function from (3) with
26as the input.5. Call the function from (3) with
Expected outputs for If Statements In Functions-2as the input.Size Estimates by Name (20 pts)
You’re going to write a function to estimate a dinosaur’s mass based on its length. The general form of the equation for doing this is:
mass <- a * length ^ b
The parameters
aandbvary by the group of dinosaurs, so you decide to create a function that lets you specify which dinosaur group you need to estimate the size of by name and then have the function automatically choose the right parameters.Create a new function
get_mass_from_length_by_name()that takes two arguments, thelengthand the name of the dinosaur group. Inside this function useif/else if/elsestatements to check to see if the name is one of the following values and if so use the associated equation to estimate the species mass:- Stegosauria:
mass = 10.95 * length ^ 2.64(Seebacher 2001) - Theropoda:
mass = 0.73 * length ^ 3.63(Seebacher 2001) - Sauropoda:
mass = 214.44 * length ^ 1.46(Seebacher 2001)
If the name is not any of these values the function should return
NA.Run the function for:
- A Stegosauria that is 10 meters long.
- A Theropoda that is 8 meters long.
- A Sauropoda that is 12 meters long.
- A Ankylosauria that is 13 meters long.
Challenge (optional): If the name is not one of values that have
aandbvalues print warning that it doesn’t know how to convert that group that includes that groups name in a message like “No known estimation for Ankylosauria”. You can use the functionwarning("your warning text")to print a warning and the functionpaste()to combine text with a value from a variablepaste("My name is", name). Doing this successfully will modify your answer to (4), which is fine.Challenge (optional): Change your function so that it uses two different equations for Stegosauria. When Stegosauria is greater than 8 meters long use the equation above. When it is less than 8 meters long use
Expected outputs for Size Estimates by Namemass = 8.5 * length ^ 2.8`. Run the function for a Stegosauria that is 6 meters long.- Stegosauria:
DNA or RNA (20 pts)
Write a function that determines if a sequence of base pairs is DNA, RNA, or if it is not possible to tell given the sequence provided. RNA has the base Uracil (
"u") instead of the base Thymine ("t"), so sequences with u’s are RNA, sequences with t’s are DNA, and sequences with neither are unknown.You can check if a string contains a character (or a longer substring) in R using the
str_detectfunction from thestringrpackage:str_detect(string, substring), which will returnTRUEifsubstringis present instring. Sostr_detect(sequence, "u")will check if the string in thesequencevariable has the baseu.Name the function
dna_or_rna()and have it takesequenceas an argument. Have the function return one of three outputs:"DNA","RNA", or"UNKNOWN". Call the function on each of the following sequences.seq1 <- "ttgaatgccttacaactgatcattacacaggcggcatgaagcaaaaatatactgtgaaccaatgcaggcg" seq2 <- "gauuauuccccacaaagggagugggauuaggagcugcaucauuuacaagagcagaauguuucaaaugcau" seq3 <- "gaaagcaagaaaaggcaggcgaggaagggaagaagggggggaaacc"Challenge (optional): Figure out how to make your function work with both upper and lower case letters, or even strings with mixed capitalization.
Expected outputs for DNA or RNACheck That Your Code Runs (10 pts)
Sometimes you think you’re code runs, but it only actually works because of something else you did previously. To make sure it actually runs you should save your work and then run it in a clean environment.
Follow these steps in RStudio to make sure your code really runs:
1. Restart R (see above) by clicking
Sessionin the menu bar and selectingRestart R: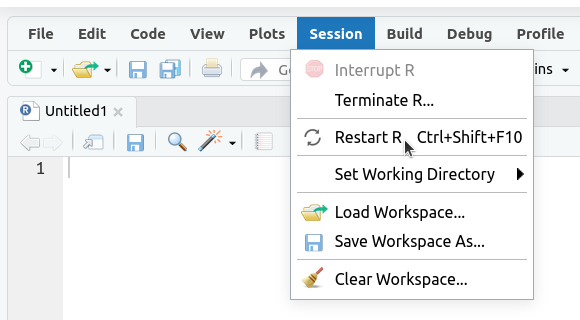
2. If the
Environmenttab isn’t empty click on the broom icon to clear it: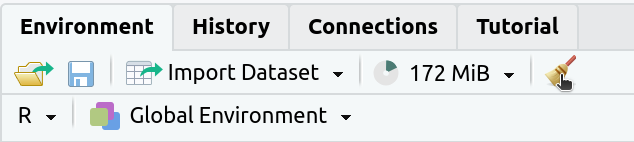
The
Environmenttab should now say “Environment Is Empty”: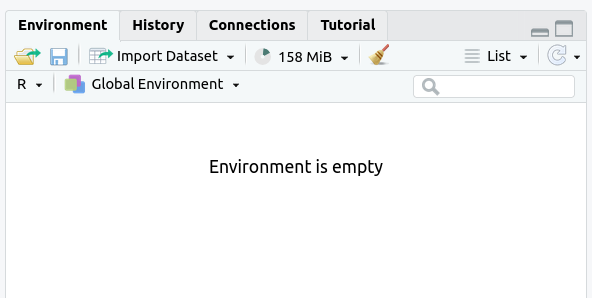
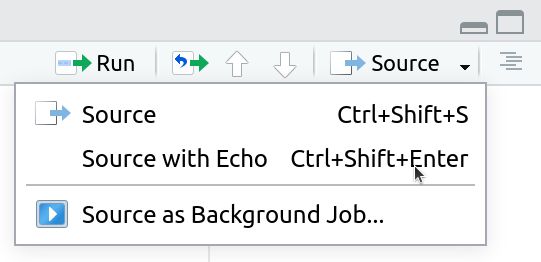
3. Rerun your entire homework assignment using “Source with Echo” to make sure it runs from start to finish and produces the expected results.
4. Make sure that you saved your code with the name
assignmentsomewhere in the file name. You should see the file in theFilestab and the name of the file should be black (not red with an*in the tab at the top of the text editor):

5. Make sure that your code will run on other computers
- No
setwd()(use RStudio Projects instead) - Use
/not\for paths
- No
Load or Download File (Challenge - optional)
With large data files it can be useful to only download the file if it hasn’t already been downloaded. One way to do this is to check if the file name exists in your working directory. If it does then load it, if not then download it. You can use the
list.files()function to get a list of files and directories in the working directory and thedownload.file(url, filename)function to download the file at aurlto a specificfilename.-
Write a conditional statement that checks if
surveys.csvexists in the working directory, if it doesn’t then downloads it from https://ndownloader.figshare.com/files/2292172 usingdownload.file(), and finally loads the file into a data frame and displays the first few rows using thehead()function. The url needs to be in quotes since it is character data. -
Make a version of this conditional statement that is a function, where the name of the file is the first argument and the link for downloading the file is the second argument. The function should return the resulting data frame. Add some documentation to the top of the function describing what it does. Call this function using “species.csv” as the file name and https://ndownloader.figshare.com/files/3299483 as the link. Print the first few rows of the resulting data frame using
head().
-
Unit Conversion Challenge (Challenge - optional)
Measures of the amount of energy used by biological processes are critical to understanding many aspects of biology from cellular physiology to ecosystem ecology. There are many different units for energy use and their utilization varies across methods, research areas, and lab groups. Write a function,
convert_energy_units(energy_value, input_unit, output_unit)to convert units between the following energy values - Joules(J), Kilojoules(KJ), Calories(CAL), and Kilocalories (KCAL; this is unit used for labeling the amount of energy contained in food). A Kilojoule is 1000 Joules, a Calorie is 4.1868 Joules, a Kilocalorie is 4186.8 Joules. An example of a call to this function would look like:energy_in_cal <- 200 energy_in_j <- convert_energy_units(energy_in_cal, "CAL", "J")Make this function more efficient by linking
if elsestatements. If either the input unit or the output unit do not match the five types given above, have the function returnNAand display a warning (using thewarning()function) - “Cannot convert:” + the name of the unit provided.Instead of writing an individual conversion between each of the different currencies (which would require 12 if statements) you could choose to convert all of the input units to a common scale and then convert from that common scale to the output units. This approach is especially useful since we might need to add new units later and this will be much easier using this approach.
Use your function to answer the following questions:
- What is the daily metabolic energy used by a human (~2500 KCALs) in Joules.
- How many times more energy does a common seal use than a human? The common seal uses ~52,500 KJ/day (Nagy et al. 1999). Use the daily human metabolic cost given above.
- How many ergs (ERG) are there in one kilocalorie. Since we didn’t include the erg conversion this should trigger our ‘Cannot convert’ message