Data Wrangling and Analyses with Tidyverse
Last updated on 2026-02-08 | Edit this page
Estimated time: 55 minutes
Overview
Questions
- How can I manipulate data frames without repeating myself?
Objectives
- Describe what the
dplyrpackage in R is used for. - Apply common
dplyrfunctions to manipulate data in R. - Employ the ‘pipe’ operator to link together a sequence of functions.
- Employ the ‘mutate’ function to apply other chosen functions to existing columns and create new columns of data.
- Employ the ‘split-apply-combine’ concept to split the data into groups, apply analysis to each group, and combine the results.
Bracket subsetting is handy, but it can be cumbersome and difficult to read, especially for complicated operations.
Luckily, the dplyr
package provides a number of very useful functions for manipulating data
frames in a way that will reduce repetition, reduce the probability of
making errors, and probably even save you some typing. As an added
bonus, you might even find the dplyr grammar easier to
read.
Here we’re going to cover some of the most commonly used functions as
well as using pipes (%>%) to combine them:
glimpse()select()filter()group_by()summarize()mutate()-
pivot_longerandpivot_wider
Packages in R are sets of additional functions that let you do more
stuff in R. The functions we’ve been using, like str(),
come built into R; packages give you access to more functions. You need
to install a package and then load it to be able to use it.
R
install.packages("dplyr") ## installs dplyr package
install.packages("tidyr") ## installs tidyr package
install.packages("ggplot2") ## installs ggplot2 package
install.packages("readr") ## install readr package
You might get asked to choose a CRAN mirror – this is asking you to choose a site to download the package from. The choice doesn’t matter too much; I’d recommend choosing the RStudio mirror.
R
library("dplyr") ## loads in dplyr package to use
library("tidyr") ## loads in tidyr package to use
library("ggplot2") ## loads in ggplot2 package to use
library("readr") ## load in readr package to use
You only need to install a package once per computer, but you need to load it every time you open a new R session and want to use that package.
Tip: Installing packages
It may be temping to install the tidyverse package, as
it contains many useful collection of packages for this lesson and
beyond. However, when teaching or following this lesson, we advise that
participants install dplyr, readr,
ggplot2, and tidyr individually as shown
above. Otherwise, a substantial amount of the lesson will be spend
waiting for the installation to complete.
What is dplyr?
The package dplyr is a fairly new (2014) package that
tries to provide easy tools for the most common data manipulation tasks.
This package is also included in the tidyverse package,
which is a collection of eight different packages (dplyr,
ggplot2, tibble, tidyr,
readr, purrr, stringr, and
forcats). It is built to work directly with data frames.
The thinking behind it was largely inspired by the package
plyr which has been in use for some time but suffered from
being slow in some cases.dplyr addresses this by porting
much of the computation to C++. An additional feature is the ability to
work with data stored directly in an external database. The benefits of
doing this are that the data can be managed natively in a relational
database, queries can be conducted on that database, and only the
results of the query returned.
This addresses a common problem with R in that all operations are conducted in memory and thus the amount of data you can work with is limited by available memory. The database connections essentially remove that limitation in that you can have a database that is over 100s of GB, conduct queries on it directly and pull back just what you need for analysis in R.
Loading .csv files in tidy style
The Tidyverse’s readr package provides its own unique
way of loading .csv files in to R using read_csv(), which
is similar to read.csv(). read_csv() allows
users to load in their data faster, doesn’t create row names, and allows
you to access non-standard variable names (i.e. variables that start
with numbers of contain spaces), and outputs your data on the R console
in a tidier way. In short, it’s a much friendlier way of loading in
potentially messy data.
Now let’s load our vcf .csv file using read_csv():
Taking a quick look at data frames
Similar to str(), which comes built into R,
glimpse() is a dplyr function that (as the
name suggests) gives a glimpse of the data frame.
OUTPUT
Rows: 801
Columns: 29
$ sample_id <chr> "SRR2584863", "SRR2584863", "SRR2584863", "SRR2584863", …
$ CHROM <chr> "CP000819.1", "CP000819.1", "CP000819.1", "CP000819.1", …
$ POS <dbl> 9972, 263235, 281923, 433359, 473901, 648692, 1331794, 1…
$ ID <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ REF <chr> "T", "G", "G", "CTTTTTTT", "CCGC", "C", "C", "G", "ACAGC…
$ ALT <chr> "G", "T", "T", "CTTTTTTTT", "CCGCGC", "T", "A", "A", "AC…
$ QUAL <dbl> 91.0000, 85.0000, 217.0000, 64.0000, 228.0000, 210.0000,…
$ FILTER <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ INDEL <lgl> FALSE, FALSE, FALSE, TRUE, TRUE, FALSE, FALSE, FALSE, TR…
$ IDV <dbl> NA, NA, NA, 12, 9, NA, NA, NA, 2, 7, NA, NA, NA, NA, NA,…
$ IMF <dbl> NA, NA, NA, 1.000000, 0.900000, NA, NA, NA, 0.666667, 1.…
$ DP <dbl> 4, 6, 10, 12, 10, 10, 8, 11, 3, 7, 9, 20, 12, 19, 15, 10…
$ VDB <dbl> 0.0257451, 0.0961330, 0.7740830, 0.4777040, 0.6595050, 0…
$ RPB <dbl> NA, 1.000000, NA, NA, NA, NA, NA, NA, NA, NA, 0.900802, …
$ MQB <dbl> NA, 1.0000000, NA, NA, NA, NA, NA, NA, NA, NA, 0.1501340…
$ BQB <dbl> NA, 1.000000, NA, NA, NA, NA, NA, NA, NA, NA, 0.750668, …
$ MQSB <dbl> NA, NA, 0.974597, 1.000000, 0.916482, 0.916482, 0.900802…
$ SGB <dbl> -0.556411, -0.590765, -0.662043, -0.676189, -0.662043, -…
$ MQ0F <dbl> 0.000000, 0.166667, 0.000000, 0.000000, 0.000000, 0.0000…
$ ICB <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ HOB <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ AC <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ AN <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ DP4 <chr> "0,0,0,4", "0,1,0,5", "0,0,4,5", "0,1,3,8", "1,0,2,7", "…
$ MQ <dbl> 60, 33, 60, 60, 60, 60, 60, 60, 60, 60, 25, 60, 10, 60, …
$ Indiv <chr> "/home/dcuser/dc_workshop/results/bam/SRR2584863.aligned…
$ gt_PL <dbl> 1210, 1120, 2470, 910, 2550, 2400, 2080, 2550, 11128, 19…
$ gt_GT <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ gt_GT_alleles <chr> "G", "T", "T", "CTTTTTTTT", "CCGCGC", "T", "A", "A", "AC…In the above output, we can already gather some information about
variants, such as the number of rows and columns, column
names, type of vector in the columns, and the first few entries of each
column. Although what we see is similar to outputs of
str(), this method gives a cleaner visual output.
Selecting columns and filtering rows
To select columns of a data frame, use select(). The
first argument to this function is the data frame
(variants), and the subsequent arguments are the columns to
keep.
R
select(variants, sample_id, REF, ALT, DP)
OUTPUT
# A tibble: 801 × 4
sample_id REF ALT DP
<chr> <chr> <chr> <dbl>
1 SRR2584863 T G 4
2 SRR2584863 G T 6
3 SRR2584863 G T 10
4 SRR2584863 CTTTTTTT CTTTTTTTT 12
5 SRR2584863 CCGC CCGCGC 10
6 SRR2584863 C T 10
7 SRR2584863 C A 8
8 SRR2584863 G A 11
9 SRR2584863 ACAGCCAGCCAGCCAGCCAGCCAGCCAGCCAG ACAGCCAGCCAGCCAGCCAGCCAGCC… 3
10 SRR2584863 AT ATT 7
# ℹ 791 more rowsTo select all columns except certain ones, put a “-” in front of the variable to exclude it.
R
select(variants, -CHROM)
OUTPUT
# A tibble: 801 × 28
sample_id POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR2584863 9972 NA T G 91 NA FALSE NA NA 4
2 SRR2584863 263235 NA G T 85 NA FALSE NA NA 6
3 SRR2584863 281923 NA G T 217 NA FALSE NA NA 10
4 SRR2584863 433359 NA CTTTTTTT CTTT… 64 NA TRUE 12 1 12
5 SRR2584863 473901 NA CCGC CCGC… 228 NA TRUE 9 0.9 10
6 SRR2584863 648692 NA C T 210 NA FALSE NA NA 10
7 SRR2584863 1331794 NA C A 178 NA FALSE NA NA 8
8 SRR2584863 1733343 NA G A 225 NA FALSE NA NA 11
9 SRR2584863 2103887 NA ACAGCCA… ACAG… 56 NA TRUE 2 0.667 3
10 SRR2584863 2333538 NA AT ATT 167 NA TRUE 7 1 7
# ℹ 791 more rows
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>dplyr also provides useful functions to select columns
based on their names. For instance, ends_with() allows you
to select columns that ends with specific letters. For instance, if you
wanted to select columns that end with the letter “B”:
R
select(variants, ends_with("B"))
OUTPUT
# A tibble: 801 × 8
VDB RPB MQB BQB MQSB SGB ICB HOB
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl> <lgl>
1 0.0257 NA NA NA NA -0.556 NA NA
2 0.0961 1 1 1 NA -0.591 NA NA
3 0.774 NA NA NA 0.975 -0.662 NA NA
4 0.478 NA NA NA 1 -0.676 NA NA
5 0.660 NA NA NA 0.916 -0.662 NA NA
6 0.268 NA NA NA 0.916 -0.670 NA NA
7 0.624 NA NA NA 0.901 -0.651 NA NA
8 0.992 NA NA NA 1.01 -0.670 NA NA
9 0.902 NA NA NA 1 -0.454 NA NA
10 0.568 NA NA NA 1.01 -0.617 NA NA
# ℹ 791 more rowsChallenge
Create a table that contains all the columns with the letter “i” and
column “POS”, without columns “Indiv” and “FILTER”. Hint: look at for a
function called contains(), which can be found in the help
documentation for ends with we just covered (?ends_with).
Note that contains() is not case sensitive.
R
# First, we select "POS" and all columns with letter "i". This will contain columns Indiv and FILTER.
variants_subset <- select(variants, POS, contains("i"))
# Next, we remove columns Indiv and FILTER
variants_result <- select(variants_subset, -Indiv, -FILTER)
variants_result
OUTPUT
# A tibble: 801 × 7
POS sample_id ID INDEL IDV IMF ICB
<dbl> <chr> <lgl> <lgl> <dbl> <dbl> <lgl>
1 9972 SRR2584863 NA FALSE NA NA NA
2 263235 SRR2584863 NA FALSE NA NA NA
3 281923 SRR2584863 NA FALSE NA NA NA
4 433359 SRR2584863 NA TRUE 12 1 NA
5 473901 SRR2584863 NA TRUE 9 0.9 NA
6 648692 SRR2584863 NA FALSE NA NA NA
7 1331794 SRR2584863 NA FALSE NA NA NA
8 1733343 SRR2584863 NA FALSE NA NA NA
9 2103887 SRR2584863 NA TRUE 2 0.667 NA
10 2333538 SRR2584863 NA TRUE 7 1 NA
# ℹ 791 more rowsChallenge (continued)
We can also get to variants_result in one line of
code:
R
variants_result <- select(variants, POS, contains("i"), -Indiv, -FILTER)
variants_result
OUTPUT
# A tibble: 801 × 7
POS sample_id ID INDEL IDV IMF ICB
<dbl> <chr> <lgl> <lgl> <dbl> <dbl> <lgl>
1 9972 SRR2584863 NA FALSE NA NA NA
2 263235 SRR2584863 NA FALSE NA NA NA
3 281923 SRR2584863 NA FALSE NA NA NA
4 433359 SRR2584863 NA TRUE 12 1 NA
5 473901 SRR2584863 NA TRUE 9 0.9 NA
6 648692 SRR2584863 NA FALSE NA NA NA
7 1331794 SRR2584863 NA FALSE NA NA NA
8 1733343 SRR2584863 NA FALSE NA NA NA
9 2103887 SRR2584863 NA TRUE 2 0.667 NA
10 2333538 SRR2584863 NA TRUE 7 1 NA
# ℹ 791 more rowsTo choose rows, use filter():
R
filter(variants, sample_id == "SRR2584863")
OUTPUT
# A tibble: 25 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl>
1 SRR2584863 CP000819… 9.97e3 NA T G 91 NA FALSE NA NA
2 SRR2584863 CP000819… 2.63e5 NA G T 85 NA FALSE NA NA
3 SRR2584863 CP000819… 2.82e5 NA G T 217 NA FALSE NA NA
4 SRR2584863 CP000819… 4.33e5 NA CTTT… CTTT… 64 NA TRUE 12 1
5 SRR2584863 CP000819… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9
6 SRR2584863 CP000819… 6.49e5 NA C T 210 NA FALSE NA NA
7 SRR2584863 CP000819… 1.33e6 NA C A 178 NA FALSE NA NA
8 SRR2584863 CP000819… 1.73e6 NA G A 225 NA FALSE NA NA
9 SRR2584863 CP000819… 2.10e6 NA ACAG… ACAG… 56 NA TRUE 2 0.667
10 SRR2584863 CP000819… 2.33e6 NA AT ATT 167 NA TRUE 7 1
# ℹ 15 more rows
# ℹ 18 more variables: DP <dbl>, VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>,
# MQSB <dbl>, SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>,
# AN <dbl>, DP4 <chr>, MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>,
# gt_GT_alleles <chr>filter() will keep all the rows that match the
conditions that are provided. Here are a few examples:
R
# rows for which the reference genome has T or G
filter(variants, REF %in% c("T", "G"))
OUTPUT
# A tibble: 340 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR25848… CP00… 9.97e3 NA T G 91 NA FALSE NA NA 4
2 SRR25848… CP00… 2.63e5 NA G T 85 NA FALSE NA NA 6
3 SRR25848… CP00… 2.82e5 NA G T 217 NA FALSE NA NA 10
4 SRR25848… CP00… 1.73e6 NA G A 225 NA FALSE NA NA 11
5 SRR25848… CP00… 2.62e6 NA G T 31.9 NA FALSE NA NA 12
6 SRR25848… CP00… 3.00e6 NA G A 225 NA FALSE NA NA 15
7 SRR25848… CP00… 3.91e6 NA G T 225 NA FALSE NA NA 10
8 SRR25848… CP00… 9.97e3 NA T G 214 NA FALSE NA NA 10
9 SRR25848… CP00… 1.06e4 NA G A 225 NA FALSE NA NA 11
10 SRR25848… CP00… 6.40e4 NA G A 225 NA FALSE NA NA 18
# ℹ 330 more rows
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>R
# rows that have TRUE in the column INDEL
filter(variants, INDEL)
OUTPUT
# A tibble: 101 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR25848… CP00… 4.33e5 NA CTTT… CTTT… 64 NA TRUE 12 1 12
2 SRR25848… CP00… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9 10
3 SRR25848… CP00… 2.10e6 NA ACAG… ACAG… 56 NA TRUE 2 0.667 3
4 SRR25848… CP00… 2.33e6 NA AT ATT 167 NA TRUE 7 1 7
5 SRR25848… CP00… 3.90e6 NA A AC 43.4 NA TRUE 2 1 2
6 SRR25848… CP00… 4.43e6 NA TGG T 228 NA TRUE 10 1 10
7 SRR25848… CP00… 1.48e5 NA AGGGG AGGG… 122 NA TRUE 8 1 8
8 SRR25848… CP00… 1.58e5 NA GTTT… GTTT… 19.5 NA TRUE 6 1 6
9 SRR25848… CP00… 1.73e5 NA CAA CA 180 NA TRUE 11 1 11
10 SRR25848… CP00… 1.75e5 NA GAA GA 194 NA TRUE 10 1 10
# ℹ 91 more rows
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>R
# rows that don't have missing data in the IDV column
filter(variants, !is.na(IDV))
OUTPUT
# A tibble: 101 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR25848… CP00… 4.33e5 NA CTTT… CTTT… 64 NA TRUE 12 1 12
2 SRR25848… CP00… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9 10
3 SRR25848… CP00… 2.10e6 NA ACAG… ACAG… 56 NA TRUE 2 0.667 3
4 SRR25848… CP00… 2.33e6 NA AT ATT 167 NA TRUE 7 1 7
5 SRR25848… CP00… 3.90e6 NA A AC 43.4 NA TRUE 2 1 2
6 SRR25848… CP00… 4.43e6 NA TGG T 228 NA TRUE 10 1 10
7 SRR25848… CP00… 1.48e5 NA AGGGG AGGG… 122 NA TRUE 8 1 8
8 SRR25848… CP00… 1.58e5 NA GTTT… GTTT… 19.5 NA TRUE 6 1 6
9 SRR25848… CP00… 1.73e5 NA CAA CA 180 NA TRUE 11 1 11
10 SRR25848… CP00… 1.75e5 NA GAA GA 194 NA TRUE 10 1 10
# ℹ 91 more rows
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>We have a column titled “QUAL”. This is a Phred-scaled confidence
score that a polymorphism exists at this position given the sequencing
data. Lower QUAL scores indicate low probability of a polymorphism
existing at that site. filter() can be useful for selecting
mutations that have a QUAL score above a certain threshold:
R
# rows with QUAL values greater than or equal to 100
filter(variants, QUAL >= 100)
OUTPUT
# A tibble: 666 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR25848… CP00… 2.82e5 NA G T 217 NA FALSE NA NA 10
2 SRR25848… CP00… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9 10
3 SRR25848… CP00… 6.49e5 NA C T 210 NA FALSE NA NA 10
4 SRR25848… CP00… 1.33e6 NA C A 178 NA FALSE NA NA 8
5 SRR25848… CP00… 1.73e6 NA G A 225 NA FALSE NA NA 11
6 SRR25848… CP00… 2.33e6 NA AT ATT 167 NA TRUE 7 1 7
7 SRR25848… CP00… 2.41e6 NA A C 104 NA FALSE NA NA 9
8 SRR25848… CP00… 2.45e6 NA A C 225 NA FALSE NA NA 20
9 SRR25848… CP00… 2.67e6 NA A T 225 NA FALSE NA NA 19
10 SRR25848… CP00… 3.00e6 NA G A 225 NA FALSE NA NA 15
# ℹ 656 more rows
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>filter() allows you to combine multiple conditions. You
can separate them using a , as arguments to the function,
they will be combined using the & (AND) logical
operator. If you need to use the | (OR) logical operator,
you can specify it explicitly:
R
# this is equivalent to:
# filter(variants, sample_id == "SRR2584863" & QUAL >= 100)
filter(variants, sample_id == "SRR2584863", QUAL >= 100)
OUTPUT
# A tibble: 19 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR25848… CP00… 2.82e5 NA G T 217 NA FALSE NA NA 10
2 SRR25848… CP00… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9 10
3 SRR25848… CP00… 6.49e5 NA C T 210 NA FALSE NA NA 10
4 SRR25848… CP00… 1.33e6 NA C A 178 NA FALSE NA NA 8
5 SRR25848… CP00… 1.73e6 NA G A 225 NA FALSE NA NA 11
6 SRR25848… CP00… 2.33e6 NA AT ATT 167 NA TRUE 7 1 7
7 SRR25848… CP00… 2.41e6 NA A C 104 NA FALSE NA NA 9
8 SRR25848… CP00… 2.45e6 NA A C 225 NA FALSE NA NA 20
9 SRR25848… CP00… 2.67e6 NA A T 225 NA FALSE NA NA 19
10 SRR25848… CP00… 3.00e6 NA G A 225 NA FALSE NA NA 15
11 SRR25848… CP00… 3.34e6 NA A C 211 NA FALSE NA NA 10
12 SRR25848… CP00… 3.40e6 NA C A 225 NA FALSE NA NA 14
13 SRR25848… CP00… 3.48e6 NA A G 200 NA FALSE NA NA 9
14 SRR25848… CP00… 3.49e6 NA A C 225 NA FALSE NA NA 13
15 SRR25848… CP00… 3.91e6 NA G T 225 NA FALSE NA NA 10
16 SRR25848… CP00… 4.10e6 NA A G 225 NA FALSE NA NA 16
17 SRR25848… CP00… 4.20e6 NA A C 225 NA FALSE NA NA 11
18 SRR25848… CP00… 4.43e6 NA TGG T 228 NA TRUE 10 1 10
19 SRR25848… CP00… 4.62e6 NA A C 185 NA FALSE NA NA 9
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>R
# using `|` logical operator
filter(variants, sample_id == "SRR2584863", (MQ >= 50 | QUAL >= 100))
OUTPUT
# A tibble: 23 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl>
1 SRR2584863 CP000819… 9.97e3 NA T G 91 NA FALSE NA NA
2 SRR2584863 CP000819… 2.82e5 NA G T 217 NA FALSE NA NA
3 SRR2584863 CP000819… 4.33e5 NA CTTT… CTTT… 64 NA TRUE 12 1
4 SRR2584863 CP000819… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9
5 SRR2584863 CP000819… 6.49e5 NA C T 210 NA FALSE NA NA
6 SRR2584863 CP000819… 1.33e6 NA C A 178 NA FALSE NA NA
7 SRR2584863 CP000819… 1.73e6 NA G A 225 NA FALSE NA NA
8 SRR2584863 CP000819… 2.10e6 NA ACAG… ACAG… 56 NA TRUE 2 0.667
9 SRR2584863 CP000819… 2.33e6 NA AT ATT 167 NA TRUE 7 1
10 SRR2584863 CP000819… 2.41e6 NA A C 104 NA FALSE NA NA
# ℹ 13 more rows
# ℹ 18 more variables: DP <dbl>, VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>,
# MQSB <dbl>, SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>,
# AN <dbl>, DP4 <chr>, MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>,
# gt_GT_alleles <chr>Challenge
Select all the mutations that occurred between the positions 1e6 (one million) and 2e6 (inclusive) that have a QUAL greater than 200, and exclude INDEL mutations. Hint: to flip logical values such as TRUE to a FALSE, we can use to negation symbol “!”. (e.g. !TRUE == FALSE).
R
filter(variants, POS >= 1e6 & POS <= 2e6, QUAL > 200, !INDEL)
OUTPUT
# A tibble: 77 × 29
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF DP
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl> <dbl>
1 SRR25848… CP00… 1.73e6 NA G A 225 NA FALSE NA NA 11
2 SRR25848… CP00… 1.00e6 NA A G 225 NA FALSE NA NA 15
3 SRR25848… CP00… 1.02e6 NA A G 225 NA FALSE NA NA 12
4 SRR25848… CP00… 1.06e6 NA C T 225 NA FALSE NA NA 17
5 SRR25848… CP00… 1.06e6 NA A G 206 NA FALSE NA NA 9
6 SRR25848… CP00… 1.07e6 NA G T 225 NA FALSE NA NA 11
7 SRR25848… CP00… 1.07e6 NA T C 225 NA FALSE NA NA 12
8 SRR25848… CP00… 1.10e6 NA C T 225 NA FALSE NA NA 15
9 SRR25848… CP00… 1.11e6 NA C T 212 NA FALSE NA NA 9
10 SRR25848… CP00… 1.11e6 NA A G 225 NA FALSE NA NA 14
# ℹ 67 more rows
# ℹ 17 more variables: VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>, MQSB <dbl>,
# SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>, AN <dbl>, DP4 <chr>,
# MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>, gt_GT_alleles <chr>Pipes
But what if you wanted to select and filter? We can do this with
pipes. Pipes, are a fairly recent addition to R. Pipes let you take the
output of one function and send it directly to the next, which is useful
when you need to many things to the same data set. It was possible to do
this before pipes were added to R, but it was much messier and more
difficult. Pipes in R look like %>% and are made
available via the magrittr package, which is installed as
part of dplyr. If you use RStudio, you can type the pipe
with Ctrl + Shift + M if you’re using a
PC, or Cmd + Shift + M if you’re using
a Mac.
R
variants %>%
filter(sample_id == "SRR2584863") %>%
select(REF, ALT, DP)
OUTPUT
# A tibble: 25 × 3
REF ALT DP
<chr> <chr> <dbl>
1 T G 4
2 G T 6
3 G T 10
4 CTTTTTTT CTTTTTTTT 12
5 CCGC CCGCGC 10
6 C T 10
7 C A 8
8 G A 11
9 ACAGCCAGCCAGCCAGCCAGCCAGCCAGCCAG ACAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGC… 3
10 AT ATT 7
# ℹ 15 more rowsIn the above code, we use the pipe to send the variants
data set first through filter(), to keep rows where
sample_id matches a particular sample, and then through
select() to keep only the REF,
ALT, and DP columns. Since %>%
takes the object on its left and passes it as the first argument to the
function on its right, we don’t need to explicitly include the data
frame as an argument to the filter() and
select() functions any more.
Some may find it helpful to read the pipe like the word “then”. For
instance, in the above example, we took the data frame
variants, then we filtered for rows
where sample_id was SRR2584863, then we
selected the REF, ALT, and
DP columns. The dplyr
functions by themselves are somewhat simple, but by combining them into
linear workflows with the pipe, we can accomplish more complex
manipulations of data frames.
If we want to create a new object with this smaller version of the data we can do so by assigning it a new name:
R
SRR2584863_variants <- variants %>%
filter(sample_id == "SRR2584863") %>%
select(REF, ALT, DP)
This new object includes all of the data from this sample. Let’s look at just the first six rows to confirm it’s what we want:
R
SRR2584863_variants
OUTPUT
# A tibble: 25 × 3
REF ALT DP
<chr> <chr> <dbl>
1 T G 4
2 G T 6
3 G T 10
4 CTTTTTTT CTTTTTTTT 12
5 CCGC CCGCGC 10
6 C T 10
7 C A 8
8 G A 11
9 ACAGCCAGCCAGCCAGCCAGCCAGCCAGCCAG ACAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGCCAGC… 3
10 AT ATT 7
# ℹ 15 more rowsSimilar to head() and tail() functions, we
can also look at the first or last six rows using tidyverse function
slice(). Slice is a more versatile function that allows
users to specify a range to view:
R
SRR2584863_variants %>% slice(1:6)
OUTPUT
# A tibble: 6 × 3
REF ALT DP
<chr> <chr> <dbl>
1 T G 4
2 G T 6
3 G T 10
4 CTTTTTTT CTTTTTTTT 12
5 CCGC CCGCGC 10
6 C T 10R
SRR2584863_variants %>% slice(10:25)
OUTPUT
# A tibble: 16 × 3
REF ALT DP
<chr> <chr> <dbl>
1 AT ATT 7
2 A C 9
3 A C 20
4 G T 12
5 A T 19
6 G A 15
7 A C 10
8 C A 14
9 A G 9
10 A C 13
11 A AC 2
12 G T 10
13 A G 16
14 A C 11
15 TGG T 10
16 A C 9Exercise: Pipe and filter
Starting with the variants data frame, use pipes to
subset the data to include only observations from SRR2584863 sample,
where the filtered depth (DP) is at least 10. Showing only 5th through
11th rows of columns REF, ALT, and
POS.
R
variants %>%
filter(sample_id == "SRR2584863" & DP >= 10) %>%
slice(5:11) %>%
select(sample_id, DP, REF, ALT, POS)
OUTPUT
# A tibble: 7 × 5
sample_id DP REF ALT POS
<chr> <dbl> <chr> <chr> <dbl>
1 SRR2584863 11 G A 1733343
2 SRR2584863 20 A C 2446984
3 SRR2584863 12 G T 2618472
4 SRR2584863 19 A T 2665639
5 SRR2584863 15 G A 2999330
6 SRR2584863 10 A C 3339313
7 SRR2584863 14 C A 3401754Mutate
Frequently you’ll want to create new columns based on the values in
existing columns, for example to do unit conversions or find the ratio
of values in two columns. For this we’ll use the dplyr
function mutate().
For example, we can convert the polymorphism confidence value QUAL to a probability value according to the formula:
Probability = 1- 10 ^ -(QUAL/10)
We can use mutate to add a column (POLPROB)
to our variants data frame that shows the probability of a
polymorphism at that site given the data.
R
variants %>%
mutate(POLPROB = 1 - (10 ^ -(QUAL/10)))
OUTPUT
# A tibble: 801 × 30
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF
<chr> <chr> <dbl> <lgl> <chr> <chr> <dbl> <lgl> <lgl> <dbl> <dbl>
1 SRR2584863 CP000819… 9.97e3 NA T G 91 NA FALSE NA NA
2 SRR2584863 CP000819… 2.63e5 NA G T 85 NA FALSE NA NA
3 SRR2584863 CP000819… 2.82e5 NA G T 217 NA FALSE NA NA
4 SRR2584863 CP000819… 4.33e5 NA CTTT… CTTT… 64 NA TRUE 12 1
5 SRR2584863 CP000819… 4.74e5 NA CCGC CCGC… 228 NA TRUE 9 0.9
6 SRR2584863 CP000819… 6.49e5 NA C T 210 NA FALSE NA NA
7 SRR2584863 CP000819… 1.33e6 NA C A 178 NA FALSE NA NA
8 SRR2584863 CP000819… 1.73e6 NA G A 225 NA FALSE NA NA
9 SRR2584863 CP000819… 2.10e6 NA ACAG… ACAG… 56 NA TRUE 2 0.667
10 SRR2584863 CP000819… 2.33e6 NA AT ATT 167 NA TRUE 7 1
# ℹ 791 more rows
# ℹ 19 more variables: DP <dbl>, VDB <dbl>, RPB <dbl>, MQB <dbl>, BQB <dbl>,
# MQSB <dbl>, SGB <dbl>, MQ0F <dbl>, ICB <lgl>, HOB <lgl>, AC <dbl>,
# AN <dbl>, DP4 <chr>, MQ <dbl>, Indiv <chr>, gt_PL <dbl>, gt_GT <dbl>,
# gt_GT_alleles <chr>, POLPROB <dbl>Exercise
There are a lot of columns in our data set, so let’s just look at the
sample_id, POS, QUAL, and
POLPROB columns for now. Add a line to the above code to
only show those columns.
R
variants %>%
mutate(POLPROB = 1 - 10 ^ -(QUAL/10)) %>%
select(sample_id, POS, QUAL, POLPROB)
OUTPUT
# A tibble: 801 × 4
sample_id POS QUAL POLPROB
<chr> <dbl> <dbl> <dbl>
1 SRR2584863 9972 91 1.000
2 SRR2584863 263235 85 1.000
3 SRR2584863 281923 217 1
4 SRR2584863 433359 64 1.000
5 SRR2584863 473901 228 1
6 SRR2584863 648692 210 1
7 SRR2584863 1331794 178 1
8 SRR2584863 1733343 225 1
9 SRR2584863 2103887 56 1.000
10 SRR2584863 2333538 167 1
# ℹ 791 more rowsgroup_by() and summarize() functions
Many data analysis tasks can be approached using the
“split-apply-combine” paradigm: split the data into groups, apply some
analysis to each group, and then combine the results. dplyr
makes this very easy through the use of the group_by()
function, which splits the data into groups.
We can use group_by() to tally the number of mutations
detected in each sample using the function tally():
R
variants %>%
group_by(sample_id) %>%
tally()
OUTPUT
# A tibble: 3 × 2
sample_id n
<chr> <int>
1 SRR2584863 25
2 SRR2584866 766
3 SRR2589044 10Since counting or tallying values is a common use case for
group_by(), an alternative function was created to bypasses
group_by() using the function count():
R
variants %>%
count(sample_id)
OUTPUT
# A tibble: 3 × 2
sample_id n
<chr> <int>
1 SRR2584863 25
2 SRR2584866 766
3 SRR2589044 10Challenge
- Count how many mutations are INDELS vs substitutions (aka not indels).
- Hint: INDELS is TRUE/FALSE column in the data.
R
variants %>%
count(INDEL)
OUTPUT
# A tibble: 2 × 2
INDEL n
<lgl> <int>
1 FALSE 700
2 TRUE 101When the data is grouped, summarize() can be used to
collapse each group into a single-row summary. summarize()
does this by applying an aggregating or summary function to each
group.
It can be a bit tricky at first, but we can imagine physically splitting the data frame by groups and applying a certain function to summarize the data.
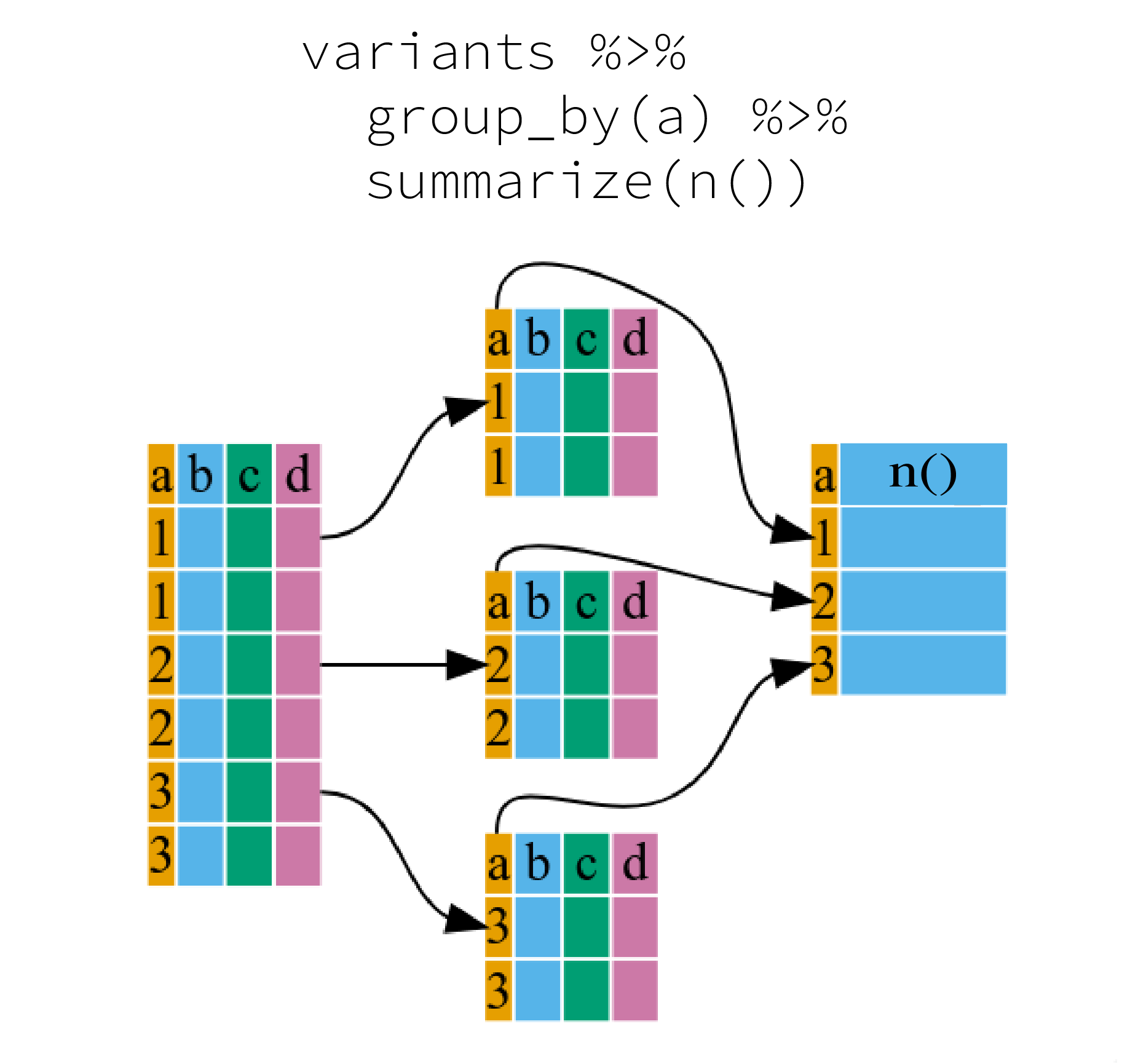
We can also apply many other functions to individual columns to get
other summary statistics. For example,we can use built-in functions like
mean(), median(), min(), and
max(). These are called “built-in functions” because they
come with R and don’t require that you install any additional packages.
By default, all R functions operating on vectors that contains
missing data will return NA. It’s a way to make sure that users
know they have missing data, and make a conscious decision on how to
deal with it. When dealing with simple statistics like the mean, the
easiest way to ignore NA (the missing data) is to use
na.rm = TRUE (rm stands for remove).
So to view the mean, median, maximum, and minimum filtered depth
(DP) for each sample:
R
variants %>%
group_by(sample_id) %>%
summarize(
mean_DP = mean(DP),
median_DP = median(DP),
min_DP = min(DP),
max_DP = max(DP))
OUTPUT
# A tibble: 3 × 5
sample_id mean_DP median_DP min_DP max_DP
<chr> <dbl> <dbl> <dbl> <dbl>
1 SRR2584863 10.4 10 2 20
2 SRR2584866 10.6 10 2 79
3 SRR2589044 9.3 9.5 3 16Grouped Data Frames in Tidyverse
When you group a data frame with group_by(), you get a
grouped data frame. This is a special type of data frame that has all
the properties of a regular data frame but also has an additional
attribute that describes the grouping structure. The primary advantage
of a grouped data frame is that it allows you to work with each group of
observations as if they were a separate data frame.
Operations like summarise() and mutate()
will be applied to each group separately. This is particularly useful
when you want to perform calculations on subsets of your data.
To remove the grouping structure from a grouped data frame, you can
use the ungroup() function. This will return a regular data
frame.
For more details, refer to the dplyr documentation on grouping.
Reshaping data frames
It can sometimes be useful to transform the “long” tidy format, into
the wide format. This transformation can be done with the
pivot_wider() function provided by the tidyr
package (also part of the tidyverse).
pivot_wider() takes a data frame as the first argument,
and two arguments: the column name that will become the columns and the
column name that will become the cells in the wide data.
R
variants_wide <- variants %>%
group_by(sample_id, CHROM) %>%
summarize(mean_DP = mean(DP)) %>%
pivot_wider(names_from = sample_id, values_from = mean_DP)
OUTPUT
`summarise()` has regrouped the output.
ℹ Summaries were computed grouped by sample_id and CHROM.
ℹ Output is grouped by sample_id.
ℹ Use `summarise(.groups = "drop_last")` to silence this message.
ℹ Use `summarise(.by = c(sample_id, CHROM))` for per-operation grouping
(`?dplyr::dplyr_by`) instead.R
variants_wide
OUTPUT
# A tibble: 1 × 4
CHROM SRR2584863 SRR2584866 SRR2589044
<chr> <dbl> <dbl> <dbl>
1 CP000819.1 10.4 10.6 9.3The opposite operation of pivot_wider() is taken care by
pivot_longer(). We specify the names of the new columns,
and here add -CHROM as this column shouldn’t be affected by
the reshaping:
R
variants_wide %>%
pivot_longer(-CHROM, names_to = "sample_id", values_to = "mean_DP")
OUTPUT
# A tibble: 3 × 3
CHROM sample_id mean_DP
<chr> <chr> <dbl>
1 CP000819.1 SRR2584863 10.4
2 CP000819.1 SRR2584866 10.6
3 CP000819.1 SRR2589044 9.3Resources
- Use the
dplyrpackage to manipulate data frames. - Use
glimpse()to quickly look at your data frame. - Use
select()to choose variables from a data frame. - Use
filter()to choose data based on values. - Use
mutate()to create new variables. - Use
group_by()andsummarize()to work with subsets of data.
The figure was adapted from the Software Carpentry lesson, R for Reproducible Scientific Analysis↩︎
