Content from Before we Start
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- How to find your way around RStudio?
- How to interact with R?
- How to manage your environment?
- How to install packages?
Objectives
- Install latest version of R.
- Install latest version of RStudio.
- Navigate the RStudio GUI.
- Install additional packages using the packages tab.
- Install additional packages using R code.
What is R? What is RStudio?
The term “R” is used to refer to both the programming
language and the software that interprets the scripts written using
it.
RStudio is currently a very popular way to not only write your R scripts but also to interact with the R software. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
To make it easier to interact with R, we will use RStudio. RStudio is the most popular IDE (Integrated Development Environment) for R. An IDE is a piece of software that provides tools to make programming easier.
You can also use the R Presentations feature to present your work in an HTML5 presentation mixing Markdown and R code. You can display these within R Studio or your browser. There are many options for customising your presentation slides, including an option for showing LaTeX equations. This can help you collaborate with others and also has an application in teaching and classroom use.
Why learn R?
R does not involve lots of pointing and clicking, and that’s a good thing
The learning curve might be steeper than with other software but with R, the results of your analysis do not rely on remembering a succession of pointing and clicking, but instead on a series of written commands, and that’s a good thing! So, if you want to redo your analysis because you collected more data, you don’t have to remember which button you clicked in which order to obtain your results; you just have to run your script again.
Working with scripts makes the steps you used in your analysis clear, and the code you write can be inspected by someone else who can give you feedback and spot mistakes.
Working with scripts forces you to have a deeper understanding of what you are doing, and facilitates your learning and comprehension of the methods you use.
R code is great for reproducibility
Reproducibility is when someone else (including your future self) can obtain the same results from the same dataset when using the same analysis.
R integrates with other tools to generate manuscripts from your code. If you collect more data, or fix a mistake in your dataset, the figures and the statistical tests in your manuscript are updated automatically.
An increasing number of journals and funding agencies expect analyses to be reproducible, so knowing R will give you an edge with these requirements.
To further support reproducibility and transparency, there are also packages that help you with dependency management: keeping track of which packages we are loading and how they depend on the package version you are using. This helps you make sure existing workflows work consistently and continue doing what they did before.
Packages like renv let you “save” and “load” the state of your project library, also keeping track of the package version you use and the source it can be retrieved from.
R is interdisciplinary and extensible
With 10,000+ packages that can be installed to extend its capabilities, R provides a framework that allows you to combine statistical approaches from many scientific disciplines to best suit the analytical framework you need to analyze your data. For instance, R has packages for image analysis, GIS, time series, population genetics, and a lot more.
R works on data of all shapes and sizes
The skills you learn with R scale easily with the size of your dataset. Whether your dataset has hundreds or millions of lines, it won’t make much difference to you.
R is designed for data analysis. It comes with special data structures and data types that make handling of missing data and statistical factors convenient.
R can connect to spreadsheets, databases, and many other data formats, on your computer or on the web.
R produces high-quality graphics
The plotting functionalities in R are endless, and allow you to adjust any aspect of your graph to convey most effectively the message from your data.
R has a large and welcoming community
Thousands of people use R daily. Many of them are willing to help you through mailing lists and websites such as Stack Overflow, or on the RStudio community. Questions which are backed up with short, reproducible code snippets are more likely to attract knowledgeable responses.
Not only is R free, but it is also open-source and cross-platform
Anyone can inspect the source code to see how R works. Because of this transparency, there is less chance for mistakes, and if you (or someone else) find some, you can report and fix bugs.
Because R is open source and is supported by a large community of developers and users, there is a very large selection of third-party add-on packages which are freely available to extend R’s native capabilities.


RStudio extends what R can do, and makes it easier to write R code and interact with R. Left photo credit; Right photo credit.
A tour of RStudio
Knowing your way around RStudio
Let’s start by learning about RStudio, which is an Integrated Development Environment (IDE) for working with R.
The RStudio IDE open-source product is free under the Affero General Public License (AGPL) v3. The RStudio IDE is also available with a commercial license and priority email support from RStudio, Inc.
We will use the RStudio IDE to write code, navigate the files on our computer, inspect the variables we create, and visualize the plots we generate. RStudio can also be used for other things (e.g., version control, developing packages, writing Shiny apps) that we will not cover during the workshop.
One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, RStudio provides many shortcuts, autocompletion, and highlighting for the major file types you use while developing in R. RStudio makes typing easier and less error-prone.
Getting set up
It is good practice to keep a set of related data, analyses, and text self-contained in a single folder called the working directory. All of the scripts within this folder can then use relative paths to files. Relative paths indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer). Working this way makes it a lot easier to move your project around on your computer and share it with others without having to directly modify file paths in the individual scripts.
RStudio provides a helpful set of tools to do this through its “Projects” interface, which not only creates a working directory for you but also remembers its location (allowing you to quickly navigate to it). The interface also (optionally) preserves custom settings and open files to make it easier to resume work after a break.
Create a new project
- Under the
Filemenu, click onNew project, chooseNew directory, thenNew project - Enter a name for this new folder (or “directory”) and choose a
convenient location for it. This will be your working
directory for the rest of the day (e.g.,
~/data-carpentry) - Click on
Create project - Create a new file where we will type our scripts. Go to File >
New File > R script. Click the save icon on your toolbar and save
your script as “
script.R”.
The simplest way to open an RStudio project once it has been created
is to navigate through your files to where the project was saved and
double click on the .Rproj (blue cube) file. This will open
RStudio and start your R session in the same directory
as the .Rproj file. All your data, plots and scripts will
now be relative to the project directory. RStudio projects have the
added benefit of allowing you to open multiple projects at the same time
each open to its own project directory. This allows you to keep multiple
projects open without them interfering with each other.
The RStudio Interface
Let’s take a quick tour of RStudio.
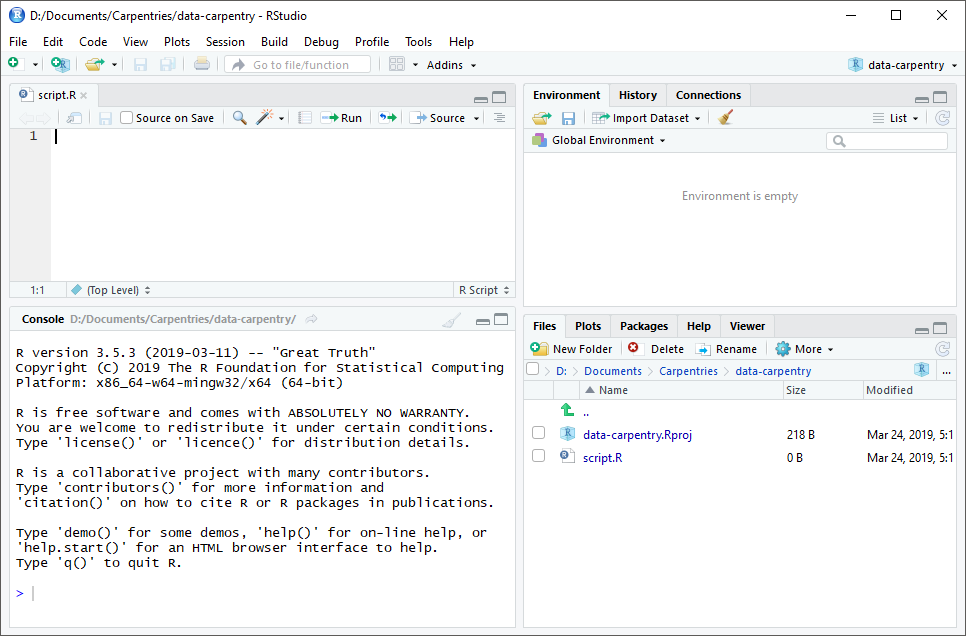
RStudio is divided into four “panes”. The placement of these panes and their content can be customized (see menu, Tools -> Global Options -> Pane Layout).
The Default Layout is:
- Top Left - Source: your scripts and documents
- Bottom Left - Console: what R would look and be like without RStudio
- Top Right - Environment/History: look here to see what you have done
- Bottom Right - Files and more: see the contents of the project/working directory here, like your Script.R file
Organizing your working directory
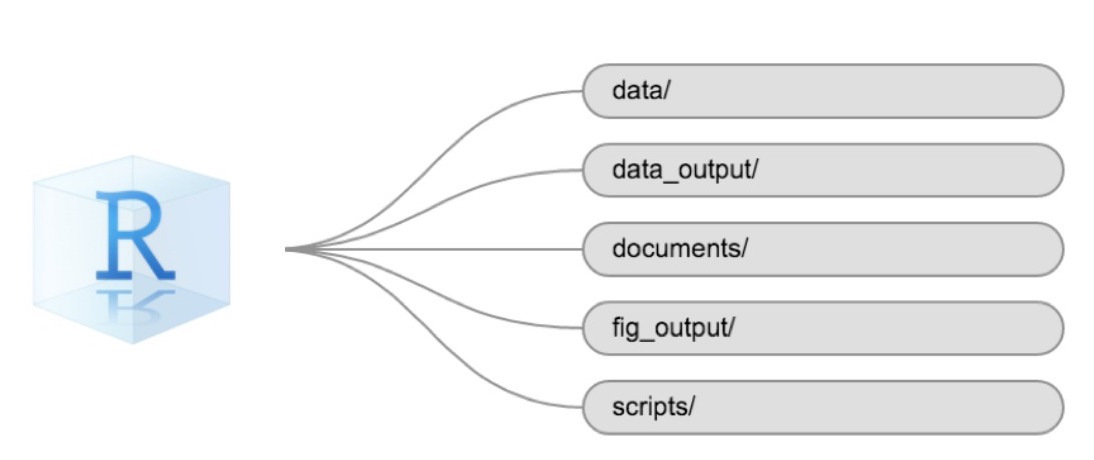
Using a consistent folder structure across your projects will help keep things organized and make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you might create directories (folders) for scripts, data, and documents. Here are some examples of suggested directories:
-
data/Use this folder to store your raw data and intermediate datasets. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually) as possible. -
data_output/When you need to modify your raw data, it might be useful to store the modified versions of the datasets in a different folder. -
documents/Used for outlines, drafts, and other text. -
fig_output/This folder can store the graphics that are generated by your scripts. -
scripts/A place to keep your R scripts for different analyses or plotting.
You may want additional directories or subdirectories depending on your project needs, but these should form the backbone of your working directory.
The working directory
The working directory is an important concept to understand. It is the place where R will look for and save files. When you write code for your project, your scripts should refer to files in relation to the root of your working directory and only to files within this structure.
Using RStudio projects makes this easy and ensures that your working
directory is set up properly. If you need to check it, you can use
getwd(). If for some reason your working directory is not
the same as the location of your RStudio project, it is likely that you
opened an R script or RMarkdown file not your
.Rproj file. You should close out of RStudio and open the
.Rproj file by double clicking on the blue cube! If you
ever need to modify your working directory in a script,
setwd('my/path') changes the working directory. This should
be used with caution since it makes analyses hard to share across
devices and with other users.
Downloading the data and getting set up
For this lesson we will use the following folders in our working
directory: data/,
data_output/ and
fig_output/. Let’s write them all in
lowercase to be consistent. We can create them using the RStudio
interface by clicking on the “New Folder” button in the file pane
(bottom right), or directly from R by typing at console:
R
dir.create("data")
dir.create("data_output")
dir.create("fig_output")
You can either download the data used for this lesson from GitHub or
with R. You can copy the data from this GitHub
link and paste it into a file called SAFI_clean.csv in
the data/ directory you just created. Or you can do this
directly from R by copying and pasting this in your terminal (your
instructor can place this chunk of code in the Etherpad):
R
download.file(
"https://raw.githubusercontent.com/datacarpentry/r-socialsci/main/episodes/data/SAFI_clean.csv",
"data/SAFI_clean.csv", mode = "wb"
)
Interacting with R
The basis of programming is that we write down instructions for the computer to follow, and then we tell the computer to follow those instructions. We write, or code, instructions in R because it is a common language that both the computer and we can understand. We call the instructions commands and we tell the computer to follow the instructions by executing (also called running) those commands.
There are two main ways of interacting with R: by using the console or by using script files (plain text files that contain your code). The console pane (in RStudio, the bottom left panel) is the place where commands written in the R language can be typed and executed immediately by the computer. It is also where the results will be shown for commands that have been executed. You can type commands directly into the console and press Enter to execute those commands, but they will be forgotten when you close the session.
Because we want our code and workflow to be reproducible, it is better to type the commands we want in the script editor and save the script. This way, there is a complete record of what we did, and anyone (including our future selves!) can easily replicate the results on their computer.
RStudio allows you to execute commands directly from the script editor by using the Ctrl + Enter shortcut (on Mac, Cmd + Return will work). The command on the current line in the script (indicated by the cursor) or all of the commands in selected text will be sent to the console and executed when you press Ctrl + Enter. If there is information in the console you do not need anymore, you can clear it with Ctrl + L. You can find other keyboard shortcuts in this RStudio cheatsheet about the RStudio IDE.
At some point in your analysis, you may want to check the content of a variable or the structure of an object without necessarily keeping a record of it in your script. You can type these commands and execute them directly in the console. RStudio provides the Ctrl + 1 and Ctrl + 2 shortcuts allow you to jump between the script and the console panes.
If R is ready to accept commands, the R console shows a
> prompt. If R receives a command (by typing,
copy-pasting, or sent from the script editor using Ctrl +
Enter), R will try to execute it and, when ready, will show
the results and come back with a new > prompt to wait
for new commands.
If R is still waiting for you to enter more text, the console will
show a + prompt. It means that you haven’t finished
entering a complete command. This is likely because you have not
‘closed’ a parenthesis or quotation, i.e. you don’t have the same number
of left-parentheses as right-parentheses or the same number of opening
and closing quotation marks. When this happens, and you thought you
finished typing your command, click inside the console window and press
Esc; this will cancel the incomplete command and return you
to the > prompt. You can then proofread the command(s)
you entered and correct the error.
Installing additional packages using the packages tab
In addition to the core R installation, there are in excess of 10,000 additional packages which can be used to extend the functionality of R. Many of these have been written by R users and have been made available in central repositories, like the one hosted at CRAN, for anyone to download and install into their own R environment. You should have already installed the packages ‘ggplot2’ and ’dplyr. If you have not, please do so now using these instructions.
You can see if you have a package installed by looking in the
packages tab (on the lower-right by default). You can also
type the command installed.packages() into the console and
examine the output.
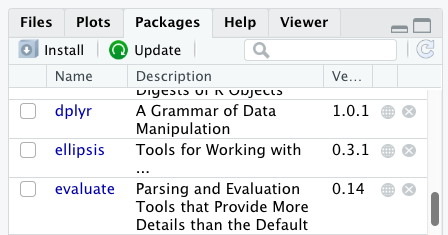
Additional packages can be installed from the ‘packages’ tab. On the packages tab, click the ‘Install’ icon and start typing the name of the package you want in the text box. As you type, packages matching your starting characters will be displayed in a drop-down list so that you can select them.
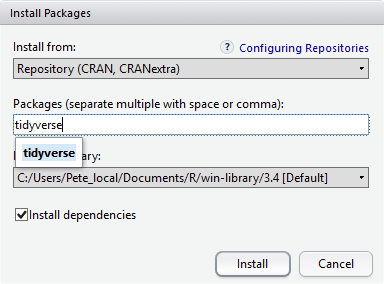
At the bottom of the Install Packages window is a check box to ‘Install’ dependencies. This is ticked by default, which is usually what you want. Packages can (and do) make use of functionality built into other packages, so for the functionality contained in the package you are installing to work properly, there may be other packages which have to be installed with them. The ‘Install dependencies’ option makes sure that this happens.
Exercise
Use both the Console and the Packages tab to confirm that you have the tidyverse installed.
Scroll through packages tab down to ‘tidyverse’. You can also type a few characters into the searchbox. The ‘tidyverse’ package is really a package of packages, including ‘ggplot2’ and ‘dplyr’, both of which require other packages to run correctly. All of these packages will be installed automatically. Depending on what packages have previously been installed in your R environment, the install of ‘tidyverse’ could be very quick or could take several minutes. As the install proceeds, messages relating to its progress will be written to the console. You will be able to see all of the packages which are actually being installed.
Because the install process accesses the CRAN repository, you will need an Internet connection to install packages.
It is also possible to install packages from other repositories, as well as Github or the local file system, but we won’t be looking at these options in this lesson.
Installing additional packages using R code
If you were watching the console window when you started the install of ‘tidyverse’, you may have noticed that the line
R
install.packages("tidyverse")
was written to the console before the start of the installation messages.
You could also have installed the
tidyverse packages by running this command
directly at the R terminal.
We will be using another package called
here throughout the workshop to manage
paths and directories. We will discuss it more detail in a later
episode, but we will install it now in the console:
R
install.packages("here")
- Use RStudio to write and run R programs.
- Use
install.packages()to install packages (libraries).
Content from Introduction to R
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- What data types are available in R?
- What is an object?
- How can objects of different data types be assigned to names?
- What arithmetic and logical operators can be used?
- How can subsets be extracted from vectors?
- How does R treat missing values?
- How can we deal with missing values in R?
Objectives
- Define the following terms as they relate to R: object, assign, call, function, arguments, options.
- Assign values to names in R.
- Learn how to name objects.
- Use comments to inform script.
- Solve simple arithmetic operations in R.
- Call functions and use arguments to change their default options.
- Inspect the content of vectors and manipulate their content.
- Subset values from vectors.
- Analyze vectors with missing data.
Creating objects in R
You can get output from R simply by typing math in the console:
R
3 + 5
OUTPUT
[1] 8R
12 / 7
OUTPUT
[1] 1.714286Everything that exists in R is an objects: from simple
numerical values, to strings, to more complex objects like vectors,
matrices, and lists. Even expressions and functions are objects in
R.
However, to do useful and interesting things, we need to name
objects. To do so, we need to give a name followed by the
assignment operator <-, and the object we want
to be named:
R
area_hectares <- 1.0
<- is the assignment operator. It assigns values
(objects) on the right to names (also called symbols) on the
left. So, after executing x <- 3, the value of
x is 3. The arrow can be read as 3
goes into x. For historical reasons, you
can also use = for assignments, but not in every context.
Because of the slight
differences in syntax, it is good practice to always use
<- for assignments. More generally we prefer the
<- syntax over = because it makes it clear
what direction the assignment is operating (left assignment), and it
increases the read-ability of the code.
In RStudio, typing Alt + - (push Alt
at the same time as the - key) will write <-
in a single keystroke in a PC, while typing Option +
- (push Option at the same time as the
- key) does the same in a Mac.
Objects can be given any name such as x,
current_temperature, or subject_id. You want
your object names to be explicit and not too long. They cannot start
with a number (2x is not valid, but x2 is). R
is case sensitive (e.g., age is different from
Age). There are some names that cannot be used because they
are the names of fundamental objects in R (e.g., if,
else, for, see here
for a complete list). In general, even if it’s allowed, it’s best to not
use them (e.g., c, T, mean,
data, df, weights). If in doubt,
check the help to see if the name is already in use. It’s also best to
avoid dots (.) within an object name as in
my.dataset. There are many objects in R with dots in their
names for historical reasons, but because dots have a special meaning in
R (for methods) and other programming languages, it’s best to avoid
them. The recommended writing style is called snake_case, which implies
using only lowercaseletters and numbers and separating each word with
underscores (e.g., animals_weight, average_income). It is also
recommended to use nouns for object names, and verbs for function names.
It’s important to be consistent in the styling of your code (where you
put spaces, how you name objects, etc.). Using a consistent coding style
makes your code clearer to read for your future self and your
collaborators. In R, three popular style guides are Google’s, Jean Fan’s and the tidyverse’s. The tidyverse’s is
very comprehensive and may seem overwhelming at first. You can install
the lintr
package to automatically check for issues in the styling of your
code.
Objects vs. variables
The naming of objects in R is somehow related to
variables in many other programming languages. In many
programming languages, a variable has three aspects: a name, a memory
location, and the current value stored in this location. R
abstracts from modifiable memory locations. In R we only
have objects which can be named. Depending on the context,
name (of an object) and variable can have
drastically different meanings. However, in this lesson, the two words
are used synonymously. For more information see: https://cran.r-project.org/doc/manuals/r-release/R-lang.html#Objects
When assigning an value to a name, R does not print anything. You can force R to print the value by using parentheses or by typing the object name:
R
area_hectares <- 1.0 # doesn't print anything
(area_hectares <- 1.0) # putting parenthesis around the call prints the value of `area_hectares`
OUTPUT
[1] 1R
area_hectares # and so does typing the name of the object
OUTPUT
[1] 1Now that R has area_hectares in memory, we can do
arithmetic with it. For instance, we may want to convert this area into
acres (area in acres is 2.47 times the area in hectares):
R
2.47 * area_hectares
OUTPUT
[1] 2.47We can also change an the value assigned to an name by assigning it a new one:
R
area_hectares <- 2.5
2.47 * area_hectares
OUTPUT
[1] 6.175This means that assigning a value to one name does not change the
values of other names. For example, let’s name the plot’s area in acres
area_acres:
R
area_acres <- 2.47 * area_hectares
and then change (reassign) area_hectares to 50.
R
area_hectares <- 50
Exercise
What do you think is the current value of area_acres?
123.5 or 6.175?
The value of area_acres is still 6.175 because you have
not re-run the line area_acres <- 2.47 * area_hectares
since changing the value of area_hectares.
Comments
All programming languages allow the programmer to include comments in their code. Including comments to your code has many advantages: it helps you explain your reasoning and it forces you to be tidy. A commented code is also a great tool not only to your collaborators, but to your future self. Comments are the key to a reproducible analysis.
To do this in R we use the # character. Anything to the
right of the # sign and up to the end of the line is
treated as a comment and is ignored by R. You can start lines with
comments or include them after any code on the line.
R
area_hectares <- 1.0 # land area in hectares
area_acres <- area_hectares * 2.47 # convert to acres
area_acres # print land area in acres.
OUTPUT
[1] 2.47RStudio makes it easy to comment or uncomment a paragraph: after selecting the lines you want to comment, press at the same time on your keyboard Ctrl + Shift + C. If you only want to comment out one line, you can put the cursor at any location of that line (i.e. no need to select the whole line), then press Ctrl + Shift + C.
Exercise
Create two variables r_length and r_width
and assign them values. It should be noted that, because
length is a built-in R function, R Studio might add “()”
after you type length and if you leave the parentheses you
will get unexpected results. This is why you might see other programmers
abbreviate common words. Create a third variable r_area and
give it a value based on the current values of r_length and
r_width. Show that changing the values of either
r_length and r_width does not affect the value
of r_area.
R
r_length <- 2.5
r_width <- 3.2
r_area <- r_length * r_width
r_area
OUTPUT
[1] 8R
# change the values of r_length and r_width
r_length <- 7.0
r_width <- 6.5
# the value of r_area isn't changed
r_area
OUTPUT
[1] 8Functions and their arguments
Functions are “canned scripts” that automate more complicated sets of
commands including operations assignments, etc. Many functions are
predefined, or can be made available by importing R packages
(more on that later). A function usually gets one or more inputs called
arguments. Functions often (but not always) return a
value. A typical example would be the function
sqrt(). The input (the argument) must be a number, and the
return value (in fact, the output) is the square root of that number.
Executing a function (‘running it’) is called calling the
function. An example of a function call is:
R
b <- sqrt(a)
Here, the value of a is given to the sqrt()
function, the sqrt() function calculates the square root,
and returns the value which is then assigned to the name b.
This function is very simple, because it takes just one argument.
The return ‘value’ of a function need not be numerical (like that of
sqrt()), and it also does not need to be a single item: it
can be a set of things, or even a dataset. We’ll see that when we read
data files into R.
Arguments can be anything, not only numbers or filenames, but also other objects. Exactly what each argument means differs per function, and must be looked up in the documentation (see below). Some functions take arguments which may either be specified by the user, or, if left out, take on a default value: these are called options. Options are typically used to alter the way the function operates, such as whether it ignores ‘bad values’, or what symbol to use in a plot. However, if you want something specific, you can specify a value of your choice which will be used instead of the default.
Let’s try a function that can take multiple arguments:
round().
R
round(3.14159)
OUTPUT
[1] 3Here, we’ve called round() with just one argument,
3.14159, and it has returned the value 3.
That’s because the default is to round to the nearest whole number. If
we want more digits we can see how to do that by getting information
about the round function. We can use
args(round) or look at the help for this function using
?round.
R
args(round)
OUTPUT
function (x, digits = 0, ...)
NULLR
?round
We see that if we want a different number of digits, we can type
digits=2 or however many we want.
R
round(3.14159, digits = 2)
OUTPUT
[1] 3.14If you provide the arguments in the exact same order as they are defined you don’t have to name them:
R
round(3.14159, 2)
OUTPUT
[1] 3.14And if you do name the arguments, you can switch their order:
R
round(digits = 2, x = 3.14159)
OUTPUT
[1] 3.14It’s good practice to put the non-optional arguments (like the number you’re rounding) first in your function call, and to specify the names of all optional arguments. If you don’t, someone reading your code might have to look up the definition of a function with unfamiliar arguments to understand what you’re doing.
Exercise
Type in ?round at the console and then look at the
output in the Help pane. What other functions exist that are similar to
round? How do you use the digits parameter in
the round function?
Vectors and data types
A vector is the most common and basic data type in R, and is pretty
much the workhorse of R. A vector is composed by a series of values,
which can be either numbers or characters. We can assign a series of
values to a vector using the c() function. For example we
can create a vector of the number of household members for the
households we’ve interviewed and assign it to
hh_members:
R
hh_members <- c(3, 7, 10, 6)
hh_members
OUTPUT
[1] 3 7 10 6A vector can also contain characters. For example, we can have a
vector of the building material used to construct our interview
respondents’ walls (respondent_wall_type):
R
respondent_wall_type <- c("muddaub", "burntbricks", "sunbricks")
respondent_wall_type
OUTPUT
[1] "muddaub" "burntbricks" "sunbricks" The quotes around “muddaub”, etc. are essential here. Without the
quotes R will assume there are objects called muddaub,
burntbricks and sunbricks. As these names
don’t exist in R’s memory, there will be an error message.
There are many functions that allow you to inspect the content of a
vector. length() tells you how many elements are in a
particular vector:
R
length(hh_members)
OUTPUT
[1] 4R
length(respondent_wall_type)
OUTPUT
[1] 3An important feature of a vector, is that all of the elements are the
same type of data. The function typeof() indicates the type
of an object:
R
typeof(hh_members)
OUTPUT
[1] "double"R
typeof(respondent_wall_type)
OUTPUT
[1] "character"The function str() provides an overview of the structure
of an object and its elements. It is a useful function when working with
large and complex objects:
R
str(hh_members)
OUTPUT
num [1:4] 3 7 10 6R
str(respondent_wall_type)
OUTPUT
chr [1:3] "muddaub" "burntbricks" "sunbricks"You can use the c() function to add other elements to
your vector:
R
possessions <- c("bicycle", "radio", "television")
possessions <- c(possessions, "mobile_phone") # add to the end of the vector
possessions <- c("car", possessions) # add to the beginning of the vector
possessions
OUTPUT
[1] "car" "bicycle" "radio" "television" "mobile_phone"In the first line, we take the original vector
possessions, add the value "mobile_phone" to
the end of it, and save the result back into possessions.
Then we add the value "car" to the beginning, again saving
the result back into possessions.
We can do this over and over again to grow a vector, or assemble a dataset. As we program, this may be useful to add results that we are collecting or calculating.
An atomic vector is the simplest R data
type and is a linear vector of a single type. Above, we saw 2
of the 6 main atomic vector types that R uses:
"character" and "numeric" (or
"double"). These are the basic building blocks that all R
objects are built from. The other 4 atomic vector types
are:
-
"logical"forTRUEandFALSE(the boolean data type) -
"integer"for integer numbers (e.g.,2L, theLindicates to R that it’s an integer) -
"complex"to represent complex numbers with real and imaginary parts (e.g.,1 + 4i) and that’s all we’re going to say about them -
"raw"for bitstreams that we won’t discuss further
You can check the type of your vector using the typeof()
function and inputting your vector as the argument.
Vectors are one of the many data structures that R
uses. Other important ones are lists (list), matrices
(matrix), data frames (data.frame), factors
(factor) and arrays (array).
Exercise
We’ve seen that atomic vectors can be of type character, numeric (or double), integer, and logical. But what happens if we try to mix these types in a single vector?
R implicitly converts them to all be the same type.
Exercise (continued)
What will happen in each of these examples? (hint: use
class() to check the data type of your objects):
R
num_char <- c(1, 2, 3, "a")
num_logical <- c(1, 2, 3, TRUE)
char_logical <- c("a", "b", "c", TRUE)
tricky <- c(1, 2, 3, "4")
Why do you think it happens?
Vectors can be of only one data type. R tries to convert (coerce) the content of this vector to find a “common denominator” that doesn’t lose any information.
Exercise (continued)
How many values in combined_logical are
"TRUE" (as a character) in the following example:
R
num_logical <- c(1, 2, 3, TRUE)
char_logical <- c("a", "b", "c", TRUE)
combined_logical <- c(num_logical, char_logical)
Only one. There is no memory of past data types, and the coercion
happens the first time the vector is evaluated. Therefore, the
TRUE in num_logical gets converted into a
1 before it gets converted into "1" in
combined_logical.
Exercise (continued)
You’ve probably noticed that objects of different types get converted into a single, shared type within a vector. In R, we call converting objects from one class into another class coercion. These conversions happen according to a hierarchy, whereby some types get preferentially coerced into other types. Can you draw a diagram that represents the hierarchy of how these data types are coerced?
Subsetting vectors
Subsetting (sometimes referred to as extracting or indexing) involves accessing out one or more values based on their numeric placement or “index” within a vector. If we want to subset one or several values from a vector, we must provide one index or several indices in square brackets. For instance:
R
respondent_wall_type <- c("muddaub", "burntbricks", "sunbricks")
respondent_wall_type[2]
OUTPUT
[1] "burntbricks"R
respondent_wall_type[c(3, 2)]
OUTPUT
[1] "sunbricks" "burntbricks"We can also repeat the indices to create an object with more elements than the original one:
R
more_respondent_wall_type <- respondent_wall_type[c(1, 2, 3, 2, 1, 3)]
more_respondent_wall_type
OUTPUT
[1] "muddaub" "burntbricks" "sunbricks" "burntbricks" "muddaub"
[6] "sunbricks" R indices start at 1. Programming languages like Fortran, MATLAB, Julia, and R start counting at 1, because that’s what human beings typically do. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that’s simpler for computers to do.
Conditional subsetting
Another common way of subsetting is by using a logical vector.
TRUE will select the element with the same index, while
FALSE will not:
R
hh_members <- c(3, 7, 10, 6)
hh_members[c(TRUE, FALSE, TRUE, TRUE)]
OUTPUT
[1] 3 10 6Typically, these logical vectors are not typed by hand, but are the output of other functions or logical tests. For instance, if you wanted to select only the values above 5:
R
hh_members > 5 # will return logicals with TRUE for the indices that meet the condition
OUTPUT
[1] FALSE TRUE TRUE TRUER
## so we can use this to select only the values above 5
hh_members[hh_members > 5]
OUTPUT
[1] 7 10 6You can combine multiple tests using & (both
conditions are true, AND) or | (at least one of the
conditions is true, OR):
R
hh_members[hh_members < 4 | hh_members > 7]
OUTPUT
[1] 3 10R
hh_members[hh_members >= 4 & hh_members <= 7]
OUTPUT
[1] 7 6Here, < stands for “less than”, > for
“greater than”, >= for “greater than or equal to”, and
== for “equal to”. The double equal sign == is
a test for numerical equality between the left and right hand sides, and
should not be confused with the single = sign, which
performs variable assignment (similar to <-).
A common task is to search for certain strings in a vector. One could
use the “or” operator | to test for equality to multiple
values, but this can quickly become tedious.
R
possessions <- c("car", "bicycle", "radio", "television", "mobile_phone")
possessions[possessions == "car" | possessions == "bicycle"] # returns both car and bicycle
OUTPUT
[1] "car" "bicycle"The function %in% allows you to test if any of the
elements of a search vector (on the left hand side) are found in the
target vector (on the right hand side):
R
possessions %in% c("car", "bicycle")
OUTPUT
[1] TRUE TRUE FALSE FALSE FALSENote that the output is the same length as the search vector on the
left hand side, because %in% checks whether each element of
the search vector is found somewhere in the target vector. Thus, you can
use %in% to select the elements in the search vector that
appear in your target vector:
R
possessions %in% c("car", "bicycle", "motorcycle", "truck", "boat", "bus")
OUTPUT
[1] TRUE TRUE FALSE FALSE FALSER
possessions[possessions %in% c("car", "bicycle", "motorcycle", "truck", "boat", "bus")]
OUTPUT
[1] "car" "bicycle"Missing data
As R was designed to analyze datasets, it includes the concept of
missing data (which is uncommon in other programming languages). Missing
data are represented in vectors as NA.
When doing operations on numbers, most functions will return
NA if the data you are working with include missing values.
This feature makes it harder to overlook the cases where you are dealing
with missing data. You can add the argument na.rm=TRUE to
calculate the result while ignoring the missing values.
R
rooms <- c(2, 1, 1, NA, 7)
mean(rooms)
OUTPUT
[1] NAR
max(rooms)
OUTPUT
[1] NAR
mean(rooms, na.rm = TRUE)
OUTPUT
[1] 2.75R
max(rooms, na.rm = TRUE)
OUTPUT
[1] 7If your data include missing values, you may want to become familiar
with the functions is.na(), na.omit(), and
complete.cases(). See below for examples.
R
## Extract those elements which are not missing values.
## The ! character is also called the NOT operator
rooms[!is.na(rooms)]
OUTPUT
[1] 2 1 1 7R
## Count the number of missing values.
## The output of is.na() is a logical vector (TRUE/FALSE equivalent to 1/0) so the sum() function here is effectively counting
sum(is.na(rooms))
OUTPUT
[1] 1R
## Returns the object with incomplete cases removed. The returned object is an atomic vector of type `"numeric"` (or `"double"`).
na.omit(rooms)
OUTPUT
[1] 2 1 1 7
attr(,"na.action")
[1] 4
attr(,"class")
[1] "omit"R
## Extract those elements which are complete cases. The returned object is an atomic vector of type `"numeric"` (or `"double"`).
rooms[complete.cases(rooms)]
OUTPUT
[1] 2 1 1 7Recall that you can use the typeof() function to find
the type of your atomic vector.
Exercise
- Using this vector of rooms, create a new vector with the NAs removed.
R
rooms <- c(1, 2, 1, 1, NA, 3, 1, 3, 2, 1, 1, 8, 3, 1, NA, 1)
Use the function
median()to calculate the median of theroomsvector.Use R to figure out how many households in the set use more than 2 rooms for sleeping.
R
rooms <- c(1, 2, 1, 1, NA, 3, 1, 3, 2, 1, 1, 8, 3, 1, NA, 1)
rooms_no_na <- rooms[!is.na(rooms)]
# or
rooms_no_na <- na.omit(rooms)
# 2.
median(rooms, na.rm = TRUE)
OUTPUT
[1] 1R
# 3.
rooms_above_2 <- rooms_no_na[rooms_no_na > 2]
length(rooms_above_2)
OUTPUT
[1] 4Now that we have learned how to write scripts, and the basics of R’s data structures, we are ready to start working with the SAFI dataset we have been using in the other lessons, and learn about data frames.
Getting help
As mentioned in the functions and their arguments
section, you can use a question mark ? to know more
about a function (for example, typing ?round).
However, there are several other ways that people often get help when they are stuck with their R code.
- Search the internet: paste the last line of your error message or
“R” and a short description of what you want to do into your favorite
search engine and you will usually find several examples where other
people have encountered the same problem and came looking for help.
- Stack Overflow can be particularly helpful for this: answers to
questions are presented as a ranked thread ordered according to how
useful other users found them to be. You can search using the
[r]tag. - Take care: copying and pasting code written by somebody else is risky unless you understand exactly what it is doing!
- Stack Overflow can be particularly helpful for this: answers to
questions are presented as a ranked thread ordered according to how
useful other users found them to be. You can search using the
- Ask somebody “in the real world”. If you have a colleague or friend with more expertise in R than you have, show them the problem you are having and ask them for help.
- Sometimes, the act of articulating your question can help you to identify what is going wrong. This is known as “rubber duck debugging” among programmers.
Generative AI
It is increasingly common for people to use generative AI chatbots such as ChatGPT to get help while coding. You will probably receive some useful guidance by presenting your error message to the chatbot and asking it what went wrong.
However, the way this help is provided by the chatbot is different. Answers on Stack Overflow have (probably) been given by a human as a direct response to the question asked. But generative AI chatbots, which are based on advanced statistical models called Large Language Models (or LLMs), respond by generating the most likely sequence of text that would follow the prompt they are given.
While responses from generative AI tools can often be helpful, they are not always reliable. These tools sometimes generate plausible but incorrect or misleading information, so (just as with an answer found on the internet) it is essential to verify their accuracy. You need the knowledge and skills to be able to understand these responses, to judge whether or not they are accurate, and to fix any errors in the code it offers you.
In addition to asking for help, programmers can use generative AI tools to generate code from scratch; extend, improve and reorganize existing code; translate code between programming languages; figure out what terms to use in a search of the internet; and more. However, there are drawbacks that you should be aware of.
The models used by these tools have been “trained” on very large volumes of data, much of it taken from the internet, and the responses they produce reflect that training data, and may recapitulate its inaccuracies or biases. The environmental costs (energy and water use) of LLMs are a lot higher than other technologies, both during development (known as training) and when an individual user uses one (also called inference). For more information see the AI Environmental Impact Primer developed by researchers at HuggingFace, an AI hosting platform. Concerns also exist about the way the data for this training was obtained, with questions raised about whether the people developing the LLMs had permission to use it. Other ethical concerns have also been raised, such as reports that workers were exploited during the training process.
We recommend that you avoid getting help from generative AI during the workshop for several reasons:
- For most problems you will encounter at this stage, help and answers can be found among the first results returned by searching the internet.
- The foundational knowledge and skills you will learn in this lesson by writing and fixing your own programs are essential to be able to evaluate the correctness and safety of any code you receive from online help or a generative AI chatbot. If you choose to use these tools in the future, the expertise you gain from learning and practicing these fundamentals on your own will help you use them more effectively.
- As you start out with programming, the mistakes you make will be the kinds that have also been made – and overcome! – by everybody else who learned to program before you. Since these mistakes and the questions you are likely to have at this stage are common, they are also better represented than other, more specialized problems and tasks in the data that was used to train generative AI tools. This means that a generative AI chatbot is more likely to produce accurate responses to questions that novices ask, which could give you a false impression of how reliable they will be when you are ready to do things that are more advanced.
- Access individual values by location using
[]. - Access arbitrary sets of data using
[c(...)]. - Use logical operations and logical vectors to access subsets of data.
Content from Starting with Data
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- What is a data.frame?
- How can I read a complete csv file into R?
- How can I get basic summary information about my dataset?
- How can I change the way R treats strings in my dataset?
- Why would I want strings to be treated differently?
- How are dates represented in R and how can I change the format?
Objectives
- Describe what a data frame is.
- Load external data from a .csv file into a data frame.
- Summarize the contents of a data frame.
- Subset values from data frames.
- Describe the difference between a factor and a string.
- Convert between strings and factors.
- Reorder and rename factors.
- Change how character strings are handled in a data frame.
- Examine and change date formats.
What are data frames?
Data frames are the de facto data structure for tabular data
in R, and what we use for data processing, statistics, and
plotting.
A data frame is the representation of data in the format of a table where the columns are vectors that all have the same length. Data frames are analogous to the more familiar spreadsheet in programs such as Excel, with one key difference. Because columns are vectors, each column must contain a single type of data (e.g., characters, integers, factors). For example, here is a figure depicting a data frame comprising a numeric, a character, and a logical vector.
Data frames can be created by hand, but most commonly they are
generated by the functions read_csv() or
read_table(); in other words, when importing spreadsheets
from your hard drive (or the web). We will now demonstrate how to import
tabular data using read_csv().
Presentation of the SAFI Data
SAFI (Studying African Farmer-Led Irrigation) is a study looking at farming and irrigation methods in Tanzania and Mozambique. The survey data was collected through interviews conducted between November 2016 and June 2017. For this lesson, we will be using a subset of the available data. For information about the full teaching dataset used in other lessons in this workshop, see the dataset description.
We will be using a subset of the cleaned version of the dataset that
was produced through cleaning in OpenRefine
(data/SAFI_clean.csv). In this dataset, the missing data is
encoded as “NULL”, each row holds information for a single interview
respondent, and the columns represent:
| column_name | description |
|---|---|
| key_id | Added to provide a unique Id for each observation. (The InstanceID field does this as well but it is not as convenient to use) |
| village | Village name |
| interview_date | Date of interview |
| no_membrs | How many members in the household? |
| years_liv | How many years have you been living in this village or neighboring village? |
| respondent_wall_type | What type of walls does their house have (from list) |
| rooms | How many rooms in the main house are used for sleeping? |
| memb_assoc | Are you a member of an irrigation association? |
| affect_conflicts | Have you been affected by conflicts with other irrigators in the area? |
| liv_count | Number of livestock owned. |
| items_owned | Which of the following items are owned by the household? (list) |
| no_meals | How many meals do people in your household normally eat in a day? |
| months_lack_food | Indicate which months, In the last 12 months have you faced a situation when you did not have enough food to feed the household? |
| instanceID | Unique identifier for the form data submission |
Importing data
You are going to load the data in R’s memory using the function
read_csv() from the readr
package, which is part of the tidyverse;
learn more about the tidyverse collection
of packages here.
readr gets installed as part as the
tidyverse installation. When you load the
tidyverse
(library(tidyverse)), the core packages (the packages used
in most data analyses) get loaded, including
readr.
Before proceeding, however, this is a good opportunity to talk about
conflicts. Certain packages we load can end up introducing function
names that are already in use by pre-loaded R packages. For instance,
when we load the tidyverse package below, we will introduce two
conflicting functions: filter() and lag().
This happens because filter and lag are
already functions used by the stats package (already pre-loaded in R).
What will happen now is that if we, for example, call the
filter() function, R will use the
dplyr::filter() version and not the
stats::filter() one. This happens because, if conflicted,
by default R uses the function from the most recently loaded package.
Conflicted functions may cause you some trouble in the future, so it is
important that we are aware of them so that we can properly handle them,
if we want.
To do so, we just need the following functions from the conflicted package:
-
conflicted::conflict_scout(): Shows us any conflicted functions.
-
conflict_prefer("function", "package_prefered"): Allows us to choose the default function we want from now on.
It is also important to know that we can, at any time, just call the
function directly from the package we want, such as
stats::filter().
Even with the use of an RStudio project, it can be difficult to learn
how to specify paths to file locations. Enter the here
package! The here package creates paths relative to the top-level
directory (your RStudio project). These relative paths work
regardless of where the associated source file lives inside
your project, like analysis projects with data and reports in different
subdirectories. This is an important contrast to using
setwd(), which depends on the way you order your files on
your computer.

Before we can use the read_csv() and here()
functions, we need to load the tidyverse and here packages.
Also, if you recall, the missing data is encoded as “NULL” in the
dataset. We’ll tell it to the function, so R will automatically convert
all the “NULL” entries in the dataset into NA.
R
library(tidyverse)
library(here)
interviews <- read_csv(
here("data", "SAFI_clean.csv"),
na = "NULL")
In the above code, we notice the here() function takes
folder and file names as inputs (e.g., "data",
"SAFI_clean.csv"), each enclosed in quotations
("") and separated by a comma. The here() will
accept as many names as are necessary to navigate to a particular file
(e.g.,
here("analysis", "data", "surveys", "clean", "SAFI_clean.csv)).
The here() function can accept the folder and file names
in an alternate format, using a slash (“/”) rather than commas to
separate the names. The two methods are equivalent, so that
here("data", "SAFI_clean.csv") and
here("data/SAFI_clean.csv") produce the same result. (The
slash is used on all operating systems; backslashes are not used.)
If you were to type in the code above, it is likely that the
read.csv() function would appear in the automatically
populated list of functions. This function is different from the
read_csv() function, as it is included in the “base”
packages that come pre-installed with R. Overall,
read.csv() behaves similar to read_csv(), with
a few notable differences. First, read.csv() coerces column
names with spaces and/or special characters to different names
(e.g. interview date becomes interview.date).
Second, read.csv() stores data as a
data.frame, where read_csv() stores data as a
different kind of data frame called a tibble. We prefer
tibbles because they have nice printing properties among other desirable
qualities. Read more about tibbles here.
The second statement in the code above creates a data frame but
doesn’t output any data because, as you might recall, assignments
(<-) don’t display anything. (Note, however, that
read_csv may show informational text about the data frame
that is created.) If we want to check that our data has been loaded, we
can see the contents of the data frame by typing its name:
interviews in the console.
R
interviews
## Try also
## view(interviews)
## head(interviews)
OUTPUT
# A tibble: 131 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 1 God 2016-11-17 00:00:00 3 4 muddaub
2 2 God 2016-11-17 00:00:00 7 9 muddaub
3 3 God 2016-11-17 00:00:00 10 15 burntbricks
4 4 God 2016-11-17 00:00:00 7 6 burntbricks
5 5 God 2016-11-17 00:00:00 7 40 burntbricks
6 6 God 2016-11-17 00:00:00 3 3 muddaub
7 7 God 2016-11-17 00:00:00 6 38 muddaub
8 8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
9 9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
10 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
# ℹ 121 more rows
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>Note
read_csv() assumes that fields are delimited by commas.
However, in several countries, the comma is used as a decimal separator
and the semicolon (;) is used as a field delimiter. If you want to read
in this type of files in R, you can use the read_csv2
function. It behaves exactly like read_csv but uses
different parameters for the decimal and the field separators. If you
are working with another format, they can be both specified by the user.
Check out the help for read_csv() by typing
?read_csv to learn more. There is also the
read_tsv() for tab-separated data files, and
read_delim() allows you to specify more details about the
structure of your file.
Note that read_csv() actually loads the data as a
tibble. A tibble is an extension of R data frames used by
the tidyverse. When the data is read using
read_csv(), it is stored in an object of class
tbl_df, tbl, and data.frame. You
can see the class of an object with
R
class(interviews)
OUTPUT
[1] "spec_tbl_df" "tbl_df" "tbl" "data.frame" As a tibble, the type of data included in each column is
listed in an abbreviated fashion below the column names. For instance,
here key_ID is a column of floating point numbers
(abbreviated <dbl> for the word ‘double’),
village is a column of characters
(<chr>) and the interview_date is a
column in the “date and time” format (<dttm>).
Inspecting data frames
When calling a tbl_df object (like
interviews here), there is already a lot of information
about our data frame being displayed such as the number of rows, the
number of columns, the names of the columns, and as we just saw the
class of data stored in each column. However, there are functions to
extract this information from data frames. Here is a non-exhaustive list
of some of these functions. Let’s try them out!
Size:
-
dim(interviews)- returns a vector with the number of rows as the first element, and the number of columns as the second element (the dimensions of the object) -
nrow(interviews)- returns the number of rows -
ncol(interviews)- returns the number of columns
Content:
-
head(interviews)- shows the first 6 rows -
tail(interviews)- shows the last 6 rows
Names:
-
names(interviews)- returns the column names (synonym ofcolnames()fordata.frameobjects)
Summary:
-
str(interviews)- structure of the object and information about the class, length and content of each column -
summary(interviews)- summary statistics for each column -
glimpse(interviews)- returns the number of columns and rows of the tibble, the names and class of each column, and previews as many values will fit on the screen. Unlike the other inspecting functions listed above,glimpse()is not a “base R” function so you need to have thedplyrortibblepackages loaded to be able to execute it.
Note: most of these functions are “generic.” They can be used on other types of objects besides data frames or tibbles.
Subsetting data frames
Our interviews data frame has rows and columns (it has 2
dimensions). In practice, we may not need the entire data frame; for
instance, we may only be interested in a subset of the observations (the
rows) or a particular set of variables (the columns). If we want to
access some specific data from it, we need to specify the “coordinates”
(i.e., indices) we want from it. Row numbers come first, followed by
column numbers.
Tip
Subsetting a tibble with [ always results
in a tibble. However, note this is not true in general for
data frames, so be careful! Different ways of specifying these
coordinates can lead to results with different classes. This is covered
in the Software Carpentry lesson R for
Reproducible Scientific Analysis.
R
## first element in the first column of the tibble
interviews[1, 1]
OUTPUT
# A tibble: 1 × 1
key_ID
<dbl>
1 1R
## first element in the 6th column of the tibble
interviews[1, 6]
OUTPUT
# A tibble: 1 × 1
respondent_wall_type
<chr>
1 muddaub R
## first column of the tibble (as a vector)
interviews[[1]]
OUTPUT
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
[19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
[37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
[55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 127
[73] 133 152 153 155 178 177 180 181 182 186 187 195 196 197 198 201 202 72
[91] 73 76 83 85 89 101 103 102 78 80 104 105 106 109 110 113 118 125
[109] 119 115 108 116 117 144 143 150 159 160 165 166 167 174 175 189 191 192
[127] 126 193 194 199 200R
## first column of the tibble
interviews[1]
OUTPUT
# A tibble: 131 × 1
key_ID
<dbl>
1 1
2 2
3 3
4 4
5 5
6 6
7 7
8 8
9 9
10 10
# ℹ 121 more rowsR
## first three elements in the 7th column of the tibble
interviews[1:3, 7]
OUTPUT
# A tibble: 3 × 1
rooms
<dbl>
1 1
2 1
3 1R
## the 3rd row of the tibble
interviews[3, ]
OUTPUT
# A tibble: 1 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 3 God 2016-11-17 00:00:00 10 15 burntbricks
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>R
## equivalent to head_interviews <- head(interviews)
head_interviews <- interviews[1:6, ]
: is a special function that creates numeric vectors of
integers in increasing or decreasing order, test 1:10 and
10:1 for instance.
You can also exclude certain indices of a data frame using the
“-” sign:
R
interviews[, -1] # The whole tibble, except the first column
OUTPUT
# A tibble: 131 × 13
village interview_date no_membrs years_liv respondent_wall_type rooms
<chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 God 2016-11-17 00:00:00 3 4 muddaub 1
2 God 2016-11-17 00:00:00 7 9 muddaub 1
3 God 2016-11-17 00:00:00 10 15 burntbricks 1
4 God 2016-11-17 00:00:00 7 6 burntbricks 1
5 God 2016-11-17 00:00:00 7 40 burntbricks 1
6 God 2016-11-17 00:00:00 3 3 muddaub 1
7 God 2016-11-17 00:00:00 6 38 muddaub 1
8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks 3
9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks 1
10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks 5
# ℹ 121 more rows
# ℹ 7 more variables: memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>R
interviews[-c(7:131), ] # Equivalent to head(interviews)
OUTPUT
# A tibble: 6 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 1 God 2016-11-17 00:00:00 3 4 muddaub
2 2 God 2016-11-17 00:00:00 7 9 muddaub
3 3 God 2016-11-17 00:00:00 10 15 burntbricks
4 4 God 2016-11-17 00:00:00 7 6 burntbricks
5 5 God 2016-11-17 00:00:00 7 40 burntbricks
6 6 God 2016-11-17 00:00:00 3 3 muddaub
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>tibbles can be subset by calling indices (as shown
previously), but also by calling their column names directly:
R
interviews["village"] # Result is a tibble
interviews[, "village"] # Result is a tibble
interviews[["village"]] # Result is a vector
interviews$village # Result is a vector
In RStudio, you can use the autocompletion feature to get the full and correct names of the columns.
Exercise
- Create a tibble (
interviews_100) containing only the data in row 100 of theinterviewsdataset.
Now, continue using interviews for each of the following
activities:
- Notice how
nrow()gave you the number of rows in the tibble?
- Use that number to pull out just that last row in the tibble.
- Compare that with what you see as the last row using
tail()to make sure it’s meeting expectations. - Pull out that last row using
nrow()instead of the row number. - Create a new tibble (
interviews_last) from that last row.
Using the number of rows in the interviews dataset that you found in question 2, extract the row that is in the middle of the dataset. Store the content of this middle row in an object named
interviews_middle. (hint: This dataset has an odd number of rows, so finding the middle is a bit trickier than dividing n_rows by 2. Use the median( ) function and what you’ve learned about sequences in R to extract the middle row!Combine
nrow()with the-notation above to reproduce the behavior ofhead(interviews), keeping just the first through 6th rows of the interviews dataset.
R
## 1.
interviews_100 <- interviews[100, ]
## 2.
# Saving `n_rows` to improve readability and reduce duplication
n_rows <- nrow(interviews)
interviews_last <- interviews[n_rows, ]
## 3.
interviews_middle <- interviews[median(1:n_rows), ]
## 4.
interviews_head <- interviews[-(7:n_rows), ]
Factors
R has a special data class, called factor, to deal with categorical data that you may encounter when creating plots or doing statistical analyses. Factors are very useful and actually contribute to making R particularly well suited to working with data. So we are going to spend a little time introducing them.
Factors represent categorical data. They are stored as integers
associated with labels and they can be ordered (ordinal) or unordered
(nominal). Factors create a structured relation between the different
levels (values) of a categorical variable, such as days of the week or
responses to a question in a survey. This can make it easier to see how
one element relates to the other elements in a column. While factors
look (and often behave) like character vectors, they are actually
treated as integer vectors by R. So you need to be very
careful when treating them as strings.
Once created, factors can only contain a pre-defined set of values, known as levels. By default, R always sorts levels in alphabetical order. For instance, if you have a factor with 2 levels:
R
respondent_floor_type <- factor(c("earth", "cement", "cement", "earth"))
R will assign 1 to the level "cement" and
2 to the level "earth" (because c
comes before e, even though the first element in this
vector is "earth"). You can see this by using the function
levels() and you can find the number of levels using
nlevels():
R
levels(respondent_floor_type)
OUTPUT
[1] "cement" "earth" R
nlevels(respondent_floor_type)
OUTPUT
[1] 2Sometimes, the order of the factors does not matter. Other times you
might want to specify the order because it is meaningful (e.g., “low”,
“medium”, “high”). It may improve your visualization, or it may be
required by a particular type of analysis. Here, one way to reorder our
levels in the respondent_floor_type vector would be:
R
respondent_floor_type # current order
OUTPUT
[1] earth cement cement earth
Levels: cement earthR
respondent_floor_type <- factor(respondent_floor_type,
levels = c("earth", "cement"))
respondent_floor_type # after re-ordering
OUTPUT
[1] earth cement cement earth
Levels: earth cementIn R’s memory, these factors are represented by integers (1, 2), but
are more informative than integers because factors are self describing:
"cement", "earth" is more descriptive than
1, and 2. Which one is “earth”? You wouldn’t
be able to tell just from the integer data. Factors, on the other hand,
have this information built in. It is particularly helpful when there
are many levels. It also makes renaming levels easier. Let’s say we made
a mistake and need to recode “cement” to “brick”. We can do this using
the fct_recode() function from the
forcats package (included in the
tidyverse) which provides some extra tools
to work with factors.
R
levels(respondent_floor_type)
OUTPUT
[1] "earth" "cement"R
respondent_floor_type <- fct_recode(respondent_floor_type, brick = "cement")
## as an alternative, we could change the "cement" level directly using the
## levels() function, but we have to remember that "cement" is the second level
# levels(respondent_floor_type)[2] <- "brick"
levels(respondent_floor_type)
OUTPUT
[1] "earth" "brick"R
respondent_floor_type
OUTPUT
[1] earth brick brick earth
Levels: earth brickSo far, your factor is unordered, like a nominal variable. R does not
know the difference between a nominal and an ordinal variable. You make
your factor an ordered factor by using the ordered=TRUE
option inside your factor function. Note how the reported levels changed
from the unordered factor above to the ordered version below. Ordered
levels use the less than sign < to denote level
ranking.
R
respondent_floor_type_ordered <- factor(respondent_floor_type,
ordered = TRUE)
respondent_floor_type_ordered # after setting as ordered factor
OUTPUT
[1] earth brick brick earth
Levels: earth < brickConverting factors
If you need to convert a factor to a character vector, you use
as.character(x).
R
as.character(respondent_floor_type)
OUTPUT
[1] "earth" "brick" "brick" "earth"Converting factors where the levels appear as numbers (such as
concentration levels, or years) to a numeric vector is a little
trickier. The as.numeric() function returns the index
values of the factor, not its levels, so it will result in an entirely
new (and unwanted in this case) set of numbers. One method to avoid this
is to convert factors to characters, and then to numbers. Another method
is to use the levels() function. Compare:
R
year_fct <- factor(c(1990, 1983, 1977, 1998, 1990))
as.numeric(year_fct) # Wrong! And there is no warning...
OUTPUT
[1] 3 2 1 4 3R
as.numeric(as.character(year_fct)) # Works...
OUTPUT
[1] 1990 1983 1977 1998 1990R
as.numeric(levels(year_fct))[year_fct] # The recommended way.
OUTPUT
[1] 1990 1983 1977 1998 1990Notice that in the recommended levels() approach, three
important steps occur:
- We obtain all the factor levels using
levels(year_fct) - We convert these levels to numeric values using
as.numeric(levels(year_fct)) - We then access these numeric values using the underlying integers of
the vector
year_fctinside the square brackets
Renaming factors
When your data is stored as a factor, you can use the
plot() function to get a quick glance at the number of
observations represented by each factor level. Let’s extract the
memb_assoc column from our data frame, convert it into a
factor, and use it to look at the number of interview respondents who
were or were not members of an irrigation association:
R
## create a vector from the data frame column "memb_assoc"
memb_assoc <- interviews$memb_assoc
## convert it into a factor
memb_assoc <- as.factor(memb_assoc)
## let's see what it looks like
memb_assoc
OUTPUT
[1] <NA> yes <NA> <NA> <NA> <NA> no yes no no <NA> yes no <NA> yes
[16] <NA> <NA> <NA> <NA> <NA> no <NA> <NA> no no no <NA> no yes <NA>
[31] <NA> yes no yes yes yes <NA> yes <NA> yes <NA> no no <NA> no
[46] no yes <NA> <NA> yes <NA> no yes no <NA> yes no no <NA> no
[61] yes <NA> <NA> <NA> no yes no no no no yes <NA> no yes <NA>
[76] <NA> yes no no yes no no yes no yes no no <NA> yes yes
[91] yes yes yes no no no no yes no no yes yes no <NA> no
[106] no <NA> no no <NA> no <NA> <NA> no no no no yes no no
[121] no no no no no no no no no yes <NA>
Levels: no yesR
## bar plot of the number of interview respondents who were
## members of irrigation association:
plot(memb_assoc)
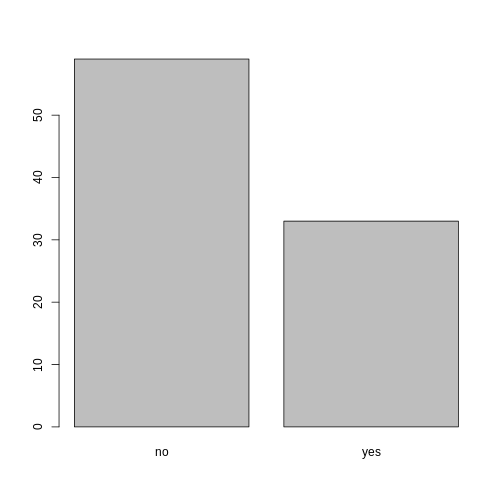
Looking at the plot compared to the output of the vector, we can see that in addition to “no”s and “yes”s, there are some respondents for whom the information about whether they were part of an irrigation association hasn’t been recorded, and encoded as missing data. These respondents do not appear on the plot. Let’s encode them differently so they can be counted and visualized in our plot.
R
## Let's recreate the vector from the data frame column "memb_assoc"
memb_assoc <- interviews$memb_assoc
## replace the missing data with "undetermined"
memb_assoc[is.na(memb_assoc)] <- "undetermined"
## convert it into a factor
memb_assoc <- as.factor(memb_assoc)
## let's see what it looks like
memb_assoc
OUTPUT
[1] undetermined yes undetermined undetermined undetermined
[6] undetermined no yes no no
[11] undetermined yes no undetermined yes
[16] undetermined undetermined undetermined undetermined undetermined
[21] no undetermined undetermined no no
[26] no undetermined no yes undetermined
[31] undetermined yes no yes yes
[36] yes undetermined yes undetermined yes
[41] undetermined no no undetermined no
[46] no yes undetermined undetermined yes
[51] undetermined no yes no undetermined
[56] yes no no undetermined no
[61] yes undetermined undetermined undetermined no
[66] yes no no no no
[71] yes undetermined no yes undetermined
[76] undetermined yes no no yes
[81] no no yes no yes
[86] no no undetermined yes yes
[91] yes yes yes no no
[96] no no yes no no
[101] yes yes no undetermined no
[106] no undetermined no no undetermined
[111] no undetermined undetermined no no
[116] no no yes no no
[121] no no no no no
[126] no no no no yes
[131] undetermined
Levels: no undetermined yesR
## bar plot of the number of interview respondents who were
## members of irrigation association:
plot(memb_assoc)
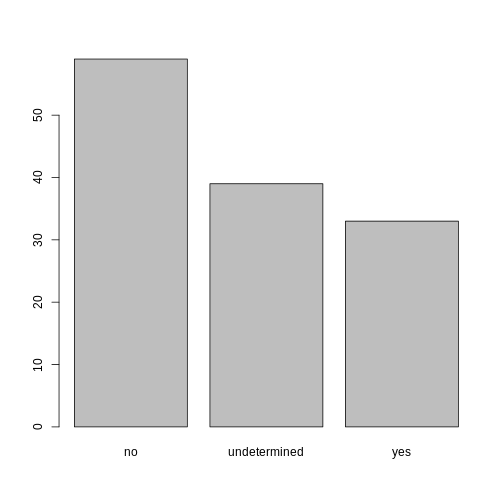
Exercise
Rename the levels of the factor to have the first letter in uppercase: “No”,“Undetermined”, and “Yes”.
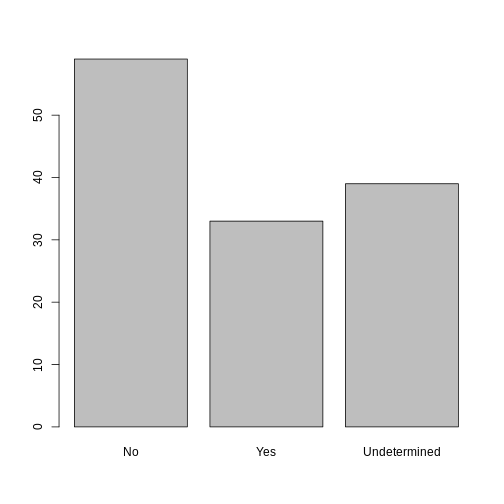
Now that we have renamed the factor level to “Undetermined”, can you recreate the barplot such that “Undetermined” is last (after “Yes”)?
R
## Rename levels.
memb_assoc <- fct_recode(memb_assoc, No = "no",
Undetermined = "undetermined", Yes = "yes")
## Reorder levels. Note we need to use the new level names.
memb_assoc <- factor(memb_assoc, levels = c("No", "Yes", "Undetermined"))
plot(memb_assoc)
Formatting Dates
One of the most common issues that new (and experienced!) R users
have is converting date and time information into a variable that is
appropriate and usable during analyses. A best practice for dealing with
date data is to ensure that each component of your date is available as
a separate variable. In our dataset, we have a column
interview_date which contains information about the year,
month, and day that the interview was conducted. Let’s convert those
dates into three separate columns.
R
str(interviews)
We are going to use the package
lubridate, , which is included in the
tidyverse installation and should be
loaded by default. However, if we deal with older versions of tidyverse
(2022 and ealier), we can manually load it by typing
library(lubridate).
If necessary, start by loading the required package:
R
library(lubridate)
The lubridate function ymd() takes a vector representing
year, month, and day, and converts it to a Date vector.
Date is a class of data recognized by R as being a date and
can be manipulated as such. The argument that the function requires is
flexible, but, as a best practice, is a character vector formatted as
“YYYY-MM-DD”.
Let’s extract our interview_date column and inspect the
structure:
R
dates <- interviews$interview_date
str(dates)
OUTPUT
POSIXct[1:131], format: "2016-11-17" "2016-11-17" "2016-11-17" "2016-11-17" "2016-11-17" ...When we imported the data in R, read_csv() recognized
that this column contained date information. We can now use the
day(), month() and year()
functions to extract this information from the date, and create new
columns in our data frame to store it:
R
interviews$day <- day(dates)
interviews$month <- month(dates)
interviews$year <- year(dates)
interviews
OUTPUT
# A tibble: 131 × 17
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 1 God 2016-11-17 00:00:00 3 4 muddaub
2 2 God 2016-11-17 00:00:00 7 9 muddaub
3 3 God 2016-11-17 00:00:00 10 15 burntbricks
4 4 God 2016-11-17 00:00:00 7 6 burntbricks
5 5 God 2016-11-17 00:00:00 7 40 burntbricks
6 6 God 2016-11-17 00:00:00 3 3 muddaub
7 7 God 2016-11-17 00:00:00 6 38 muddaub
8 8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
9 9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
10 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
# ℹ 121 more rows
# ℹ 11 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>, day <int>, month <dbl>, year <dbl>Notice the three new columns at the end of our data frame.
In our example above, the interview_date column was read
in correctly as a Date variable but generally that is not
the case. Date columns are often read in as character
variables and one can use the as_date() function to convert
them to the appropriate Date/POSIXctformat.
Let’s say we have a vector of dates in character format:
R
char_dates <- c("7/31/2012", "8/9/2014", "4/30/2016")
str(char_dates)
OUTPUT
chr [1:3] "7/31/2012" "8/9/2014" "4/30/2016"We can convert this vector to dates as :
R
as_date(char_dates, format = "%m/%d/%Y")
OUTPUT
[1] "2012-07-31" "2014-08-09" "2016-04-30"Argument format tells the function the order to parse
the characters and identify the month, day and year. The format above is
the equivalent of mm/dd/yyyy. A wrong format can lead to parsing errors
or incorrect results.
For example, observe what happens when we use a lower case y instead of upper case Y for the year.
R
as_date(char_dates, format = "%m/%d/%y")
WARNING
Warning: 3 failed to parse.OUTPUT
[1] NA NA NAHere, the %y part of the format stands for a two-digit
year instead of a four-digit year, and this leads to parsing errors.
Or in the following example, observe what happens when the month and day elements of the format are switched.
R
as_date(char_dates, format = "%d/%m/%y")
WARNING
Warning: 3 failed to parse.OUTPUT
[1] NA NA NASince there is no month numbered 30 or 31, the first and third dates cannot be parsed.
We can also use functions ymd(), mdy() or
dmy() to convert character variables to date.
R
mdy(char_dates)
OUTPUT
[1] "2012-07-31" "2014-08-09" "2016-04-30"- Use read_csv to read tabular data in R.
- Use factors to represent categorical data in R.
Content from Data Wrangling with dplyr
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- How can I select specific rows and/or columns from a dataframe?
- How can I combine multiple commands into a single command?
- How can I create new columns or remove existing columns from a dataframe?
Objectives
- Describe the purpose of an R package and the
dplyrpackage. - Select certain columns in a dataframe with the
dplyrfunctionselect. - Select certain rows in a dataframe according to filtering conditions
with the
dplyrfunctionfilter. - Link the output of one
dplyrfunction to the input of another function with the ‘pipe’ operator%>%. - Add new columns to a dataframe that are functions of existing
columns with
mutate. - Use the split-apply-combine concept for data analysis.
- Use
summarize,group_by, andcountto split a dataframe into groups of observations, apply a summary statistics for each group, and then combine the results.
dplyr is a package for making tabular
data wrangling easier by using a limited set of functions that can be
combined to extract and summarize insights from your data.
Like readr,
dplyr is a part of the tidyverse. These
packages were loaded in R’s memory when we called
library(tidyverse) earlier.
Note
The packages in the tidyverse, namely
dplyr, tidyr
and ggplot2 accept both the British
(e.g. summarise) and American (e.g. summarize)
spelling variants of different function and option names. For this
lesson, we utilize the American spellings of different functions;
however, feel free to use the regional variant for where you are
teaching.
What is an R package?
The package dplyr provides easy tools
for the most common data wrangling tasks. It is built to work directly
with dataframes, with many common tasks optimized by being written in a
compiled language (C++) (not all R packages are written in R!).
There are also packages available for a wide range of tasks including
building plots (ggplot2, which we’ll see
later), downloading data from the NCBI database, or performing
statistical analysis on your data set. Many packages such as these are
housed on, and downloadable from, the Comprehensive
R Archive Network
(CRAN) using install.packages. This function makes the
package accessible by your R installation with the command
library(), as you did with tidyverse
earlier.
To easily access the documentation for a package within R or RStudio,
use help(package = "package_name").
To learn more about dplyr after the
workshop, you may want to check out this handy
data transformation with dplyr
cheatsheet.
Note
There are alternatives to the tidyverse packages for
data wrangling, including the package data.table.
See this comparison
for example to get a sense of the differences between using
base, tidyverse, and
data.table.
Learning dplyr
To make sure everyone will use the same dataset for this lesson, we’ll read again the SAFI dataset that we downloaded earlier.
R
## load the tidyverse
library(tidyverse)
library(here)
interviews <- read_csv(here("data", "SAFI_clean.csv"), na = "NULL")
## inspect the data
interviews
## preview the data
# view(interviews)
We’re going to learn some of the most common
dplyr functions:
-
select(): subset columns -
filter(): subset rows on conditions -
mutate(): create new columns by using information from other columns -
group_by()andsummarize(): create summary statistics on grouped data -
arrange(): sort results -
count(): count discrete values
Selecting columns and filtering rows
To select columns of a dataframe, use select(). The
first argument to this function is the dataframe
(interviews), and the subsequent arguments are the columns
to keep, separated by commas. Alternatively, if you are selecting
columns adjacent to each other, you can use a : to select a
range of columns, read as “select columns from ___ to ___.” You may have
done something similar in the past using subsetting.
select() is essentially doing the same thing as subsetting,
using a package (dplyr) instead of R’s base functions.
R
# to select columns throughout the dataframe
select(interviews, village, no_membrs, months_lack_food)
# to do the same thing with subsetting
interviews[c("village","no_membrs","months_lack_food")]
# to select a series of connected columns
select(interviews, village:respondent_wall_type)
To choose rows based on specific criteria, we can use the
filter() function. The argument after the dataframe is the
condition we want our final dataframe to adhere to (e.g. village name is
Chirodzo):
R
# filters observations where village name is "Chirodzo"
filter(interviews, village == "Chirodzo")
OUTPUT
# A tibble: 39 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
2 9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
3 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
4 34 Chirodzo 2016-11-17 00:00:00 8 18 burntbricks
5 35 Chirodzo 2016-11-17 00:00:00 5 45 muddaub
6 36 Chirodzo 2016-11-17 00:00:00 6 23 sunbricks
7 37 Chirodzo 2016-11-17 00:00:00 3 8 burntbricks
8 43 Chirodzo 2016-11-17 00:00:00 7 29 muddaub
9 44 Chirodzo 2016-11-17 00:00:00 2 6 muddaub
10 45 Chirodzo 2016-11-17 00:00:00 9 7 muddaub
# ℹ 29 more rows
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>You may also have noticed that the output from these call doesn’t run
off the screen anymore. It’s one of the advantages of
tbl_df (also called tibble), the central data class in the
tidyverse, compared to normal dataframes in R.
We can also specify multiple conditions within the
filter() function. We can combine conditions using either
“and” or “or” statements. In an “and” statement, an observation (row)
must meet every criteria to be included in the
resulting dataframe. To form “and” statements within dplyr, we can pass
our desired conditions as arguments in the filter()
function, separated by commas:
R
# filters observations with "and" operator (comma)
# output dataframe satisfies ALL specified conditions
filter(interviews, village == "Chirodzo",
rooms > 1,
no_meals > 2)
OUTPUT
# A tibble: 10 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
2 49 Chirodzo 2016-11-16 00:00:00 6 26 burntbricks
3 52 Chirodzo 2016-11-16 00:00:00 11 15 burntbricks
4 56 Chirodzo 2016-11-16 00:00:00 12 23 burntbricks
5 65 Chirodzo 2016-11-16 00:00:00 8 20 burntbricks
6 66 Chirodzo 2016-11-16 00:00:00 10 37 burntbricks
7 67 Chirodzo 2016-11-16 00:00:00 5 31 burntbricks
8 68 Chirodzo 2016-11-16 00:00:00 8 52 burntbricks
9 199 Chirodzo 2017-06-04 00:00:00 7 17 burntbricks
10 200 Chirodzo 2017-06-04 00:00:00 8 20 burntbricks
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>We can also form “and” statements with the &
operator instead of commas:
R
# filters observations with "&" logical operator
# output dataframe satisfies ALL specified conditions
filter(interviews, village == "Chirodzo" &
rooms > 1 &
no_meals > 2)
OUTPUT
# A tibble: 10 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
2 49 Chirodzo 2016-11-16 00:00:00 6 26 burntbricks
3 52 Chirodzo 2016-11-16 00:00:00 11 15 burntbricks
4 56 Chirodzo 2016-11-16 00:00:00 12 23 burntbricks
5 65 Chirodzo 2016-11-16 00:00:00 8 20 burntbricks
6 66 Chirodzo 2016-11-16 00:00:00 10 37 burntbricks
7 67 Chirodzo 2016-11-16 00:00:00 5 31 burntbricks
8 68 Chirodzo 2016-11-16 00:00:00 8 52 burntbricks
9 199 Chirodzo 2017-06-04 00:00:00 7 17 burntbricks
10 200 Chirodzo 2017-06-04 00:00:00 8 20 burntbricks
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>In an “or” statement, observations must meet at least one of the specified conditions. To form “or” statements we use the logical operator for “or,” which is the vertical bar (|):
R
# filters observations with "|" logical operator
# output dataframe satisfies AT LEAST ONE of the specified conditions
filter(interviews, village == "Chirodzo" | village == "Ruaca")
OUTPUT
# A tibble: 88 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
2 9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
3 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
4 23 Ruaca 2016-11-21 00:00:00 10 20 burntbricks
5 24 Ruaca 2016-11-21 00:00:00 6 4 burntbricks
6 25 Ruaca 2016-11-21 00:00:00 11 6 burntbricks
7 26 Ruaca 2016-11-21 00:00:00 3 20 burntbricks
8 27 Ruaca 2016-11-21 00:00:00 7 36 burntbricks
9 28 Ruaca 2016-11-21 00:00:00 2 2 muddaub
10 29 Ruaca 2016-11-21 00:00:00 7 10 burntbricks
# ℹ 78 more rows
# ℹ 8 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>Pipes
What if you want to select and filter at the same time? There are three ways to do this: use intermediate steps, nested functions, or pipes.
With intermediate steps, you create a temporary dataframe and use that as input to the next function, like this:
R
interviews2 <- filter(interviews, village == "Chirodzo")
interviews_ch <- select(interviews2, village:respondent_wall_type)
This is readable, but can clutter up your workspace with lots of objects that you have to name individually. With multiple steps, that can be hard to keep track of.
You can also nest functions (i.e. one function inside of another), like this:
R
interviews_ch <- select(filter(interviews, village == "Chirodzo"),
village:respondent_wall_type)
This is handy, but can be difficult to read if too many functions are nested, as R evaluates the expression from the inside out (in this case, filtering, then selecting).
The last option, pipes, are a recent addition to R. Pipes
let you take the output of one function and send it directly to the
next, which is useful when you need to do many things to the same
dataset. There are two Pipes in R: 1) %>% (called
magrittr pipe; made available via the
magrittr package, installed automatically
with dplyr) or 2) |>
(called native R pipe and it comes preinstalled with R v4.1.0 onwards).
Both the pipes are, by and large, function similarly with a few
differences (For more information, check: https://www.tidyverse.org/blog/2023/04/base-vs-magrittr-pipe/).
The choice of which pipe to be used can be changed in the Global
settings in R studio and once that is done, you can type the pipe
with:
- Ctrl + Shift + M if you have a PC or Cmd + Shift + M if you have a Mac.
R
# the following example is run using magrittr pipe but the output will be same with the native pipe
interviews %>%
filter(village == "Chirodzo") %>%
select(village:respondent_wall_type)
OUTPUT
# A tibble: 39 × 5
village interview_date no_membrs years_liv respondent_wall_type
<chr> <dttm> <dbl> <dbl> <chr>
1 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
2 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
3 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
4 Chirodzo 2016-11-17 00:00:00 8 18 burntbricks
5 Chirodzo 2016-11-17 00:00:00 5 45 muddaub
6 Chirodzo 2016-11-17 00:00:00 6 23 sunbricks
7 Chirodzo 2016-11-17 00:00:00 3 8 burntbricks
8 Chirodzo 2016-11-17 00:00:00 7 29 muddaub
9 Chirodzo 2016-11-17 00:00:00 2 6 muddaub
10 Chirodzo 2016-11-17 00:00:00 9 7 muddaub
# ℹ 29 more rowsR
#interviews |>
# filter(village == "Chirodzo") |>
# select(village:respondent_wall_type)
In the above code, we use the pipe to send the
interviews dataset first through filter() to
keep rows where village is “Chirodzo”, then through
select() to keep only the columns from village
to respondent_wall_type. Since %>% takes
the object on its left and passes it as the first argument to the
function on its right, we don’t need to explicitly include the dataframe
as an argument to the filter() and select()
functions any more.
Some may find it helpful to read the pipe like the word “then”. For
instance, in the above example, we take the dataframe
interviews, then we filter for rows
with village == "Chirodzo", then we
select columns village:respondent_wall_type.
The dplyr functions by themselves are
somewhat simple, but by combining them into linear workflows with the
pipe, we can accomplish more complex data wrangling operations.
If we want to create a new object with this smaller version of the data, we can assign it a new name:
R
interviews_ch <- interviews %>%
filter(village == "Chirodzo") %>%
select(village:respondent_wall_type)
interviews_ch
OUTPUT
# A tibble: 39 × 5
village interview_date no_membrs years_liv respondent_wall_type
<chr> <dttm> <dbl> <dbl> <chr>
1 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
2 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
3 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
4 Chirodzo 2016-11-17 00:00:00 8 18 burntbricks
5 Chirodzo 2016-11-17 00:00:00 5 45 muddaub
6 Chirodzo 2016-11-17 00:00:00 6 23 sunbricks
7 Chirodzo 2016-11-17 00:00:00 3 8 burntbricks
8 Chirodzo 2016-11-17 00:00:00 7 29 muddaub
9 Chirodzo 2016-11-17 00:00:00 2 6 muddaub
10 Chirodzo 2016-11-17 00:00:00 9 7 muddaub
# ℹ 29 more rowsNote that the final dataframe (interviews_ch) is the
leftmost part of this expression.
Exercise
Using pipes, subset the interviews data to include
interviews where respondents were members of an irrigation association
(memb_assoc) and retain only the columns
affect_conflicts, liv_count, and
no_meals.
R
interviews %>%
filter(memb_assoc == "yes") %>%
select(affect_conflicts, liv_count, no_meals)
OUTPUT
# A tibble: 33 × 3
affect_conflicts liv_count no_meals
<chr> <dbl> <dbl>
1 once 3 2
2 never 2 2
3 never 2 3
4 once 3 2
5 frequently 1 3
6 more_once 5 2
7 more_once 3 2
8 more_once 2 3
9 once 3 3
10 never 3 3
# ℹ 23 more rowsMutate
Frequently you’ll want to create new columns based on the values in
existing columns, for example to do unit conversions, or to find the
ratio of values in two columns. For this we’ll use
mutate().
We might be interested in the ratio of number of household members to rooms used for sleeping (i.e. avg number of people per room):
R
interviews %>%
mutate(people_per_room = no_membrs / rooms)
OUTPUT
# A tibble: 131 × 15
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 1 God 2016-11-17 00:00:00 3 4 muddaub
2 2 God 2016-11-17 00:00:00 7 9 muddaub
3 3 God 2016-11-17 00:00:00 10 15 burntbricks
4 4 God 2016-11-17 00:00:00 7 6 burntbricks
5 5 God 2016-11-17 00:00:00 7 40 burntbricks
6 6 God 2016-11-17 00:00:00 3 3 muddaub
7 7 God 2016-11-17 00:00:00 6 38 muddaub
8 8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
9 9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
10 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
# ℹ 121 more rows
# ℹ 9 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>, people_per_room <dbl>We may be interested in investigating whether being a member of an irrigation association had any effect on the ratio of household members to rooms. To look at this relationship, we will first remove data from our dataset where the respondent didn’t answer the question of whether they were a member of an irrigation association. These cases are recorded as “NULL” in the dataset.
To remove these cases, we could insert a filter() in the
chain:
R
interviews %>%
filter(!is.na(memb_assoc)) %>%
mutate(people_per_room = no_membrs / rooms)
OUTPUT
# A tibble: 92 × 15
key_ID village interview_date no_membrs years_liv respondent_wall_type
<dbl> <chr> <dttm> <dbl> <dbl> <chr>
1 2 God 2016-11-17 00:00:00 7 9 muddaub
2 7 God 2016-11-17 00:00:00 6 38 muddaub
3 8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks
4 9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks
5 10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks
6 12 God 2016-11-21 00:00:00 7 20 burntbricks
7 13 God 2016-11-21 00:00:00 6 8 burntbricks
8 15 God 2016-11-21 00:00:00 5 30 sunbricks
9 21 God 2016-11-21 00:00:00 8 20 burntbricks
10 24 Ruaca 2016-11-21 00:00:00 6 4 burntbricks
# ℹ 82 more rows
# ℹ 9 more variables: rooms <dbl>, memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>, people_per_room <dbl>The ! symbol negates the result of the
is.na() function. Thus, if is.na() returns a
value of TRUE (because the memb_assoc is
missing), the ! symbol negates this and says we only want
values of FALSE, where memb_assoc is
not missing.
Exercise
Create a new dataframe from the interviews data that
meets the following criteria: contains only the village
column and a new column called total_meals containing a
value that is equal to the total number of meals served in the household
per day on average (no_membrs times no_meals).
Only the rows where total_meals is greater than 20 should
be shown in the final dataframe.
Hint: think about how the commands should be ordered to produce this data frame!
R
interviews_total_meals <- interviews %>%
mutate(total_meals = no_membrs * no_meals) %>%
filter(total_meals > 20) %>%
select(village, total_meals)
Split-apply-combine data analysis and the summarize() function
Many data analysis tasks can be approached using the
split-apply-combine paradigm: split the data into groups, apply
some analysis to each group, and then combine the results.
dplyr makes this very easy through the use
of the group_by() function.
The summarize() function
group_by() is often used together with
summarize(), which collapses each group into a single-row
summary of that group. group_by() takes as arguments the
column names that contain the categorical variables for
which you want to calculate the summary statistics. So to compute the
average household size by village:
R
interviews %>%
group_by(village) %>%
summarize(mean_no_membrs = mean(no_membrs))
OUTPUT
# A tibble: 3 × 2
village mean_no_membrs
<chr> <dbl>
1 Chirodzo 7.08
2 God 6.86
3 Ruaca 7.57You can also group by multiple columns:
R
interviews %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs))
OUTPUT
`summarise()` has grouped output by 'village'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 9 × 3
# Groups: village [3]
village memb_assoc mean_no_membrs
<chr> <chr> <dbl>
1 Chirodzo no 8.06
2 Chirodzo yes 7.82
3 Chirodzo <NA> 5.08
4 God no 7.13
5 God yes 8
6 God <NA> 6
7 Ruaca no 7.18
8 Ruaca yes 9.5
9 Ruaca <NA> 6.22Note that the output is a grouped tibble of nine rows by three
columns which is indicated by the by two first lines with the
#. To obtain an ungrouped tibble, use the
ungroup function:
R
interviews %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs)) %>%
ungroup()
OUTPUT
`summarise()` has grouped output by 'village'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 9 × 3
village memb_assoc mean_no_membrs
<chr> <chr> <dbl>
1 Chirodzo no 8.06
2 Chirodzo yes 7.82
3 Chirodzo <NA> 5.08
4 God no 7.13
5 God yes 8
6 God <NA> 6
7 Ruaca no 7.18
8 Ruaca yes 9.5
9 Ruaca <NA> 6.22Notice that the second line with the # that previously
indicated the grouping has disappeared and we now only have a 9x3-tibble
without grouping. When grouping both by village and
membr_assoc, we see rows in our table for respondents who
did not specify whether they were a member of an irrigation association.
We can exclude those data from our table using a filter step.
R
interviews %>%
filter(!is.na(memb_assoc)) %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs))
OUTPUT
`summarise()` has grouped output by 'village'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 6 × 3
# Groups: village [3]
village memb_assoc mean_no_membrs
<chr> <chr> <dbl>
1 Chirodzo no 8.06
2 Chirodzo yes 7.82
3 God no 7.13
4 God yes 8
5 Ruaca no 7.18
6 Ruaca yes 9.5 Once the data are grouped, you can also summarize multiple variables at the same time (and not necessarily on the same variable). For instance, we could add a column indicating the minimum household size for each village for each group (members of an irrigation association vs not):
R
interviews %>%
filter(!is.na(memb_assoc)) %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs),
min_membrs = min(no_membrs))
OUTPUT
`summarise()` has grouped output by 'village'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 6 × 4
# Groups: village [3]
village memb_assoc mean_no_membrs min_membrs
<chr> <chr> <dbl> <dbl>
1 Chirodzo no 8.06 4
2 Chirodzo yes 7.82 2
3 God no 7.13 3
4 God yes 8 5
5 Ruaca no 7.18 2
6 Ruaca yes 9.5 5It is sometimes useful to rearrange the result of a query to inspect
the values. For instance, we can sort on min_membrs to put
the group with the smallest household first:
R
interviews %>%
filter(!is.na(memb_assoc)) %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs),
min_membrs = min(no_membrs)) %>%
arrange(min_membrs)
OUTPUT
`summarise()` has grouped output by 'village'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 6 × 4
# Groups: village [3]
village memb_assoc mean_no_membrs min_membrs
<chr> <chr> <dbl> <dbl>
1 Chirodzo yes 7.82 2
2 Ruaca no 7.18 2
3 God no 7.13 3
4 Chirodzo no 8.06 4
5 God yes 8 5
6 Ruaca yes 9.5 5To sort in descending order, we need to add the desc()
function. If we want to sort the results by decreasing order of minimum
household size:
R
interviews %>%
filter(!is.na(memb_assoc)) %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs),
min_membrs = min(no_membrs)) %>%
arrange(desc(min_membrs))
OUTPUT
`summarise()` has grouped output by 'village'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 6 × 4
# Groups: village [3]
village memb_assoc mean_no_membrs min_membrs
<chr> <chr> <dbl> <dbl>
1 God yes 8 5
2 Ruaca yes 9.5 5
3 Chirodzo no 8.06 4
4 God no 7.13 3
5 Chirodzo yes 7.82 2
6 Ruaca no 7.18 2Counting
When working with data, we often want to know the number of
observations found for each factor or combination of factors. For this
task, dplyr provides count().
For example, if we wanted to count the number of rows of data for each
village, we would do:
R
interviews %>%
count(village)
OUTPUT
# A tibble: 3 × 2
village n
<chr> <int>
1 Chirodzo 39
2 God 43
3 Ruaca 49For convenience, count() provides the sort
argument to get results in decreasing order:
R
interviews %>%
count(village, sort = TRUE)
OUTPUT
# A tibble: 3 × 2
village n
<chr> <int>
1 Ruaca 49
2 God 43
3 Chirodzo 39Exercise
How many households in the survey have an average of two meals per day? Three meals per day? Are there any other numbers of meals represented?
R
interviews %>%
count(no_meals)
OUTPUT
# A tibble: 2 × 2
no_meals n
<dbl> <int>
1 2 52
2 3 79Exercise (continued)
Use group_by() and summarize() to find the
mean, min, and max number of household members for each village. Also
add the number of observations (hint: see ?n).
R
interviews %>%
group_by(village) %>%
summarize(
mean_no_membrs = mean(no_membrs),
min_no_membrs = min(no_membrs),
max_no_membrs = max(no_membrs),
n = n()
)
OUTPUT
# A tibble: 3 × 5
village mean_no_membrs min_no_membrs max_no_membrs n
<chr> <dbl> <dbl> <dbl> <int>
1 Chirodzo 7.08 2 12 39
2 God 6.86 3 15 43
3 Ruaca 7.57 2 19 49Exercise (continued)
What was the largest household interviewed in each month?
R
# if not already included, add month, year, and day columns
library(lubridate) # load lubridate if not already loaded
interviews %>%
mutate(month = month(interview_date),
day = day(interview_date),
year = year(interview_date)) %>%
group_by(year, month) %>%
summarize(max_no_membrs = max(no_membrs))
OUTPUT
`summarise()` has grouped output by 'year'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 5 × 3
# Groups: year [2]
year month max_no_membrs
<dbl> <dbl> <dbl>
1 2016 11 19
2 2016 12 12
3 2017 4 17
4 2017 5 15
5 2017 6 15- Use the
dplyrpackage to manipulate dataframes. - Use
select()to choose variables from a dataframe. - Use
filter()to choose data based on values. - Use
group_by()andsummarize()to work with subsets of data. - Use
mutate()to create new variables.
Content from Data Wrangling with tidyr
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- How can I reformat a data frame to meet my needs?
Objectives
- Describe the concept of a wide and a long table format and for which purpose those formats are useful.
- Describe the roles of variable names and their associated values when a table is reshaped.
- Reshape a dataframe from long to wide format and back with the
pivot_widerandpivot_longercommands from thetidyrpackage. - Export a dataframe to a csv file.
dplyr pairs nicely with
tidyr which enables you to swiftly convert
between different data formats (long vs. wide) for plotting and
analysis. To learn more about tidyr after
the workshop, you may want to check out this handy
data tidying with tidyr
cheatsheet.
To make sure everyone will use the same dataset for this lesson, we’ll read again the SAFI dataset that we downloaded earlier.
R
## load the tidyverse
library(tidyverse)
library(here)
interviews <- read_csv(here("data", "SAFI_clean.csv"), na = "NULL")
## inspect the data
interviews
## preview the data
# view(interviews)
Reshaping with pivot_wider() and pivot_longer()
There are essentially three rules that define a “tidy” dataset:
- Each variable has its own column
- Each observation has its own row
- Each value must have its own cell
This graphic visually represents the three rules that define a “tidy” dataset:
 R for Data Science,
Wickham H and Grolemund G (https://r4ds.had.co.nz/index.html)
© Wickham, Grolemund 2017 This image is licenced under
Attribution-NonCommercial-NoDerivs 3.0 United States (CC-BY-NC-ND 3.0
US)
R for Data Science,
Wickham H and Grolemund G (https://r4ds.had.co.nz/index.html)
© Wickham, Grolemund 2017 This image is licenced under
Attribution-NonCommercial-NoDerivs 3.0 United States (CC-BY-NC-ND 3.0
US)
In this section we will explore how these rules are linked to the
different data formats researchers are often interested in: “wide” and
“long”. This tutorial will help you efficiently transform your data
shape regardless of original format. First we will explore qualities of
the interviews data and how they relate to these different
types of data formats.
Long and wide data formats
In the interviews data, each row contains the values of
variables associated with each record collected (each interview in the
villages). It is stated that the key_ID was “added to
provide a unique Id for each observation” and the
instanceID “does this as well but it is not as convenient
to use.”
Once we have established that key_ID and
instanceID are both unique we can use either variable as an
identifier corresponding to the 131 interview records.
R
interviews %>%
select(key_ID) %>%
distinct() %>%
nrow()
OUTPUT
[1] 131As seen in the code below, for each interview date in each village no
instanceIDs are the same. Thus, this format is what is
called a “long” data format, where each observation occupies only one
row in the dataframe.
R
interviews %>%
filter(village == "Chirodzo") %>%
select(key_ID, village, interview_date, instanceID) %>%
sample_n(size = 10)
OUTPUT
# A tibble: 10 × 4
key_ID village interview_date instanceID
<dbl> <chr> <dttm> <chr>
1 37 Chirodzo 2016-11-17 00:00:00 uuid:408c6c93-d723-45ef-8dee-1b1bd3fe20cd
2 49 Chirodzo 2016-11-16 00:00:00 uuid:2303ebc1-2b3c-475a-8916-b322ebf18440
3 64 Chirodzo 2016-11-16 00:00:00 uuid:28cfd718-bf62-4d90-8100-55fafbe45d06
4 192 Chirodzo 2017-06-03 00:00:00 uuid:f94409a6-e461-4e4c-a6fb-0072d3d58b00
5 53 Chirodzo 2016-11-16 00:00:00 uuid:cc7f75c5-d13e-43f3-97e5-4f4c03cb4b12
6 67 Chirodzo 2016-11-16 00:00:00 uuid:6c15d667-2860-47e3-a5e7-7f679271e419
7 66 Chirodzo 2016-11-16 00:00:00 uuid:a457eab8-971b-4417-a971-2e55b8702816
8 200 Chirodzo 2017-06-04 00:00:00 uuid:aa77a0d7-7142-41c8-b494-483a5b68d8a7
9 36 Chirodzo 2016-11-17 00:00:00 uuid:c90eade0-1148-4a12-8c0e-6387a36f45b1
10 50 Chirodzo 2016-11-16 00:00:00 uuid:4267c33c-53a7-46d9-8bd6-b96f58a4f92cWe notice that the layout or format of the interviews
data is in a format that adheres to rules 1-3, where
- each column is a variable
- each row is an observation
- each value has its own cell
This is called a “long” data format. But, we notice that each column represents a different variable. In the “longest” data format there would only be three columns, one for the id variable, one for the observed variable, and one for the observed value (of that variable). This data format is quite unsightly and difficult to work with, so you will rarely see it in use.
Alternatively, in a “wide” data format we see modifications to rule 1, where each column no longer represents a single variable. Instead, columns can represent different levels/values of a variable. For instance, in some data you encounter the researchers may have chosen for every survey date to be a different column.
These may sound like dramatically different data layouts, but there are some tools that make transitions between these layouts much simpler than you might think! The gif below shows how these two formats relate to each other, and gives you an idea of how we can use R to shift from one format to the other.
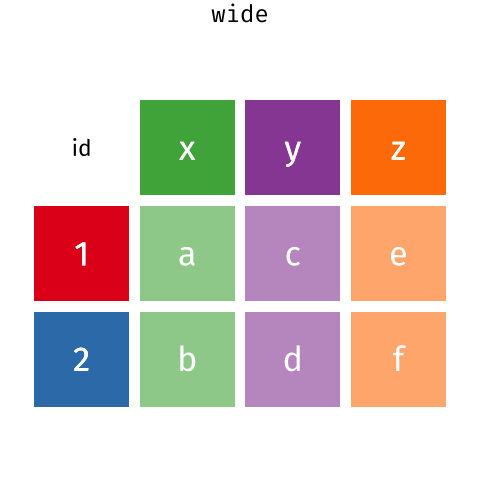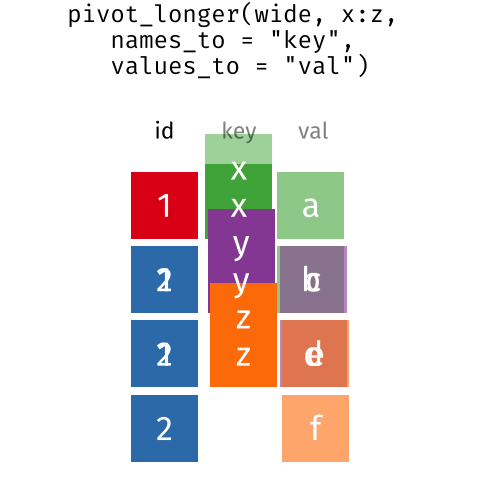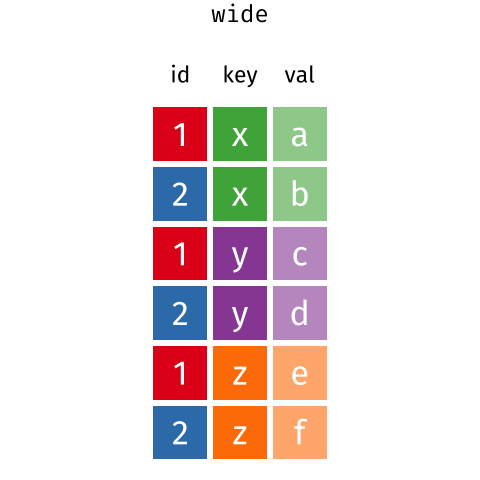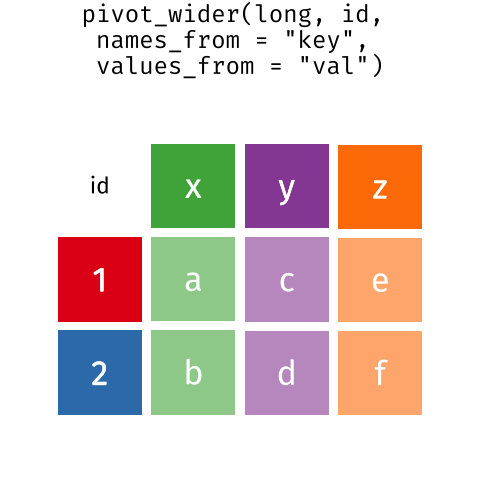 Long and wide
dataframe layouts mainly affect readability. You may find that visually
you may prefer the “wide” format, since you can see more of the data on
the screen. However, all of the R functions we have used thus far expect
for your data to be in a “long” data format. This is because the long
format is more machine readable and is closer to the formatting of
databases.
Long and wide
dataframe layouts mainly affect readability. You may find that visually
you may prefer the “wide” format, since you can see more of the data on
the screen. However, all of the R functions we have used thus far expect
for your data to be in a “long” data format. This is because the long
format is more machine readable and is closer to the formatting of
databases.
Questions which warrant different data formats
In interviews, each row contains the values of variables associated with each record (the unit), values such as the village of the respondent, the number of household members, or the type of wall their house had. This format allows for us to make comparisons across individual surveys, but what if we wanted to look at differences in households grouped by different types of items owned?
To facilitate this comparison we would need to create a new table
where each row (the unit) was comprised of values of variables
associated with items owned (i.e., items_owned). In
practical terms this means the values of the items in
items_owned (e.g. bicycle, radio, table, etc.) would become
the names of column variables and the cells would contain values of
TRUE or FALSE, for whether that household had
that item.
Once we we’ve created this new table, we can explore the relationship within and between villages. The key point here is that we are still following a tidy data structure, but we have reshaped the data according to the observations of interest.
Alternatively, if the interview dates were spread across multiple columns, and we were interested in visualizing, within each village, how irrigation conflicts have changed over time. This would require for the interview date to be included in a single column rather than spread across multiple columns. Thus, we would need to transform the column names into values of a variable.
We can do both of these transformations with two tidyr
functions, pivot_wider() and
pivot_longer().
Pivoting wider
pivot_wider() takes three principal arguments:
- the data
- the names_from column variable whose values will become new column names.
- the values_from column variable whose values will fill the new column variables.
Further arguments include values_fill which, if set,
fills in missing values with the value provided.
Let’s use pivot_wider() to transform interviews to
create new columns for each item owned by a household. There are a
couple of new concepts in this transformation, so let’s walk through it
line by line. First we create a new object
(interviews_items_owned) based on the
interviews data frame.
Then we will actually need to make our data frame longer, because we
have multiple items in a single cell. We will use a new function,
separate_longer_delim(), from the
tidyr package to separate the values of
items_owned based on the presence of semi-colons
(;). The values of this variable were multiple items
separated by semi-colons, so this action creates a row for each item
listed in a household’s possession. Thus, we end up with a long format
version of the dataset, with multiple rows for each respondent. For
example, if a respondent has a television and a solar panel, that
respondent will now have two rows, one with “television” and the other
with “solar panel” in the items_owned column.
After this transformation, you may notice that the
items_owned column contains NA values. This is
because some of the respondents did not own any of the items in the
interviewer’s list. We can use the replace_na() function to
change these NA values to something more meaningful. The
replace_na() function expects for you to give it a
list() of columns that you would like to replace the
NA values in, and the value that you would like to replace
the NAs. This ends up looking like this:
Next, we create a new variable named
items_owned_logical, which has one value
(TRUE) for every row. This makes sense, since each item in
every row was owned by that household. We are constructing this variable
so that when we spread the items_owned across multiple
columns, we can fill the values of those columns with logical values
describing whether the household did (TRUE) or did not
(FALSE) own that particular item.
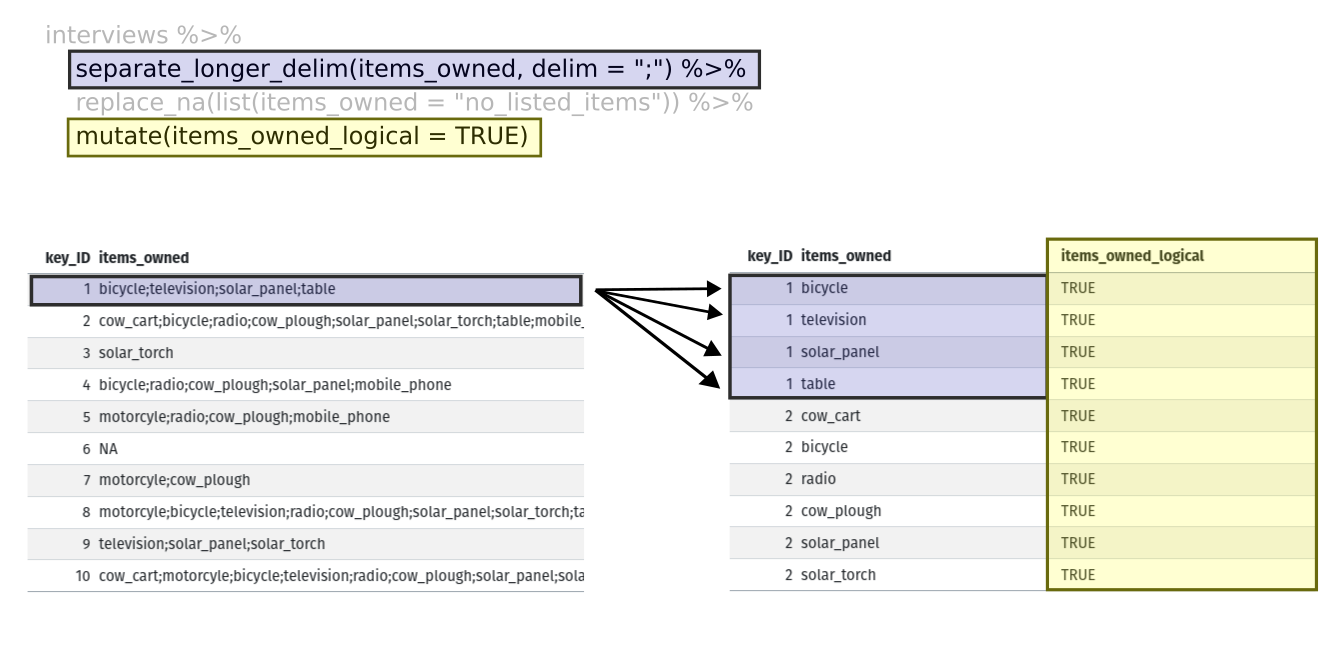
At this point, we can also count the number of items owned by each
household, which is equivalent to the number of rows per
key_ID. We can do this with a group_by() and
mutate() pipeline that works similar to
group_by() and summarize() discussed in the
previous episode but instead of creating a summary table, we will add
another column called number_items. We use the
n() function to count the number of rows within each group.
However, there is one difficulty we need to take into account, namely
those households that did not list any items. These households now have
"no_listed_items" under items_owned. We do not
want to count this as an item but instead show zero items. We can
accomplish this using dplyr’s
if_else() function that evaluates a condition and returns
one value if true and another if false. Here, if the
items_owned column is "no_listed_items", then
a 0 is returned, otherwise, the number of rows per group is returned
using n().
Lastly, we use pivot_wider() to switch from long format
to wide format. This creates a new column for each of the unique values
in the items_owned column, and fills those columns with the
values of items_owned_logical. We also declare that for
items that are missing, we want to fill those cells with the value of
FALSE instead of NA.
R
pivot_wider(names_from = items_owned,
values_from = items_owned_logical,
values_fill = list(items_owned_logical = FALSE))
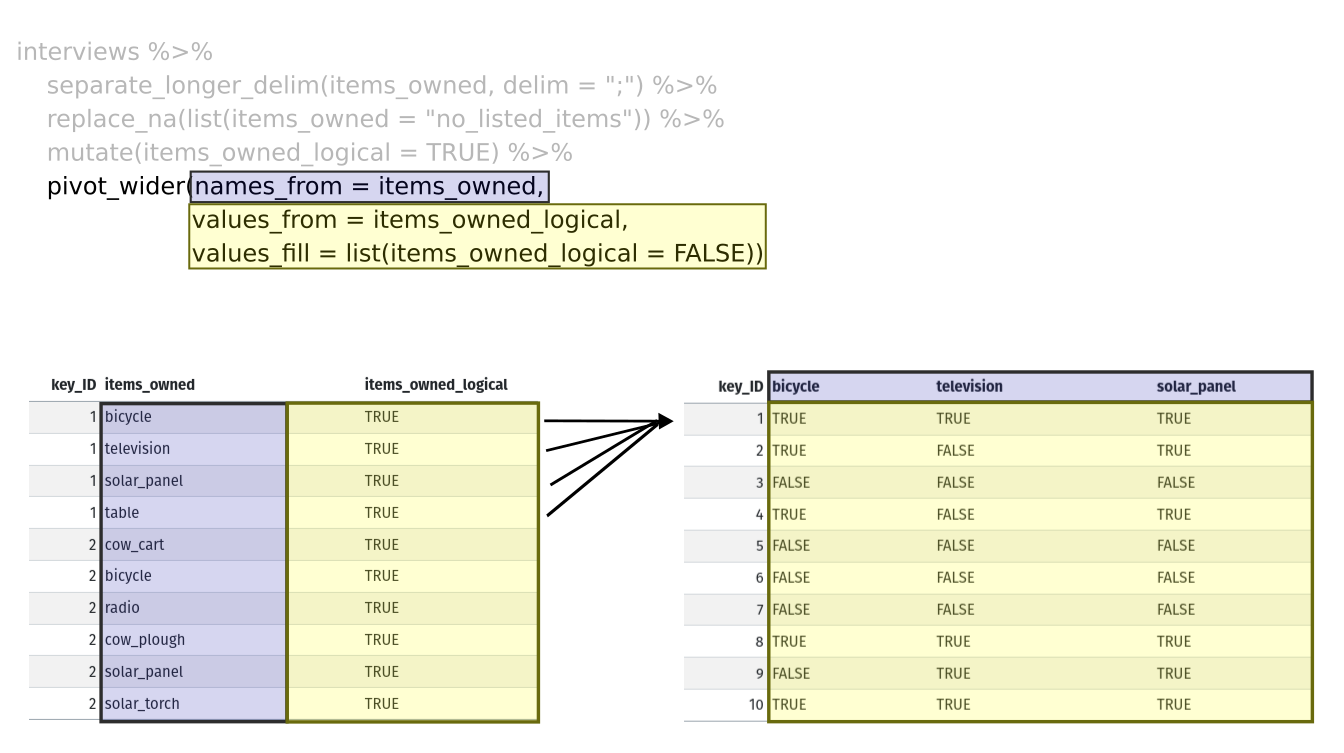
Combining the above steps, the chunk looks like this. Note that two
new columns are created within the same mutate() call.
R
interviews_items_owned <- interviews %>%
separate_longer_delim(items_owned, delim = ";") %>%
replace_na(list(items_owned = "no_listed_items")) %>%
group_by(key_ID) %>%
mutate(items_owned_logical = TRUE,
number_items = if_else(items_owned == "no_listed_items", 0, n())) %>%
pivot_wider(names_from = items_owned,
values_from = items_owned_logical,
values_fill = list(items_owned_logical = FALSE))
View the interviews_items_owned data frame. It should
have r nrow(interviews) rows (the same number of rows you
had originally), but extra columns for each item. How many columns were
added? Notice that there is no longer a column titled
items_owned. This is because there is a default parameter
in pivot_wider() that drops the original column. The values
that were in that column have now become columns named
television, solar_panel, table,
etc. You can use dim(interviews) and
dim(interviews_wide) to see how the number of columns has
changed between the two datasets.
This format of the data allows us to do interesting things, like make a table showing the number of respondents in each village who owned a particular item:
R
interviews_items_owned %>%
filter(bicycle) %>%
group_by(village) %>%
count(bicycle)
OUTPUT
# A tibble: 3 × 3
# Groups: village [3]
village bicycle n
<chr> <lgl> <int>
1 Chirodzo TRUE 17
2 God TRUE 23
3 Ruaca TRUE 20Or below we calculate the average number of items from the list owned
by respondents in each village using the number_items
column we created to count the items listed by each household.
R
interviews_items_owned %>%
group_by(village) %>%
summarize(mean_items = mean(number_items))
OUTPUT
# A tibble: 3 × 2
village mean_items
<chr> <dbl>
1 Chirodzo 4.54
2 God 3.98
3 Ruaca 5.57Exercise
We created interviews_items_owned by reshaping the data:
first longer and then wider. Replicate this process with the
months_lack_food column in the interviews
dataframe. Create a new dataframe with columns for each of the months
filled with logical vectors (TRUE or FALSE)
and a summary column called number_months_lack_food that
calculates the number of months each household reported a lack of
food.
Note that if the household did not lack food in the previous 12 months, the value input was “none”.
R
months_lack_food <- interviews %>%
separate_longer_delim(months_lack_food, delim = ";") %>%
group_by(key_ID) %>%
mutate(months_lack_food_logical = TRUE,
number_months_lack_food = if_else(months_lack_food == "none", 0, n())) %>%
pivot_wider(names_from = months_lack_food,
values_from = months_lack_food_logical,
values_fill = list(months_lack_food_logical = FALSE))
Pivoting longer
The opposing situation could occur if we had been provided with data
in the form of interviews_wide, where the items owned are
column names, but we wish to treat them as values of an
items_owned variable instead.
In this situation we are gathering these columns turning them into a pair of new variables. One variable includes the column names as values, and the other variable contains the values in each cell previously associated with the column names. We will do this in two steps to make this process a bit clearer.
pivot_longer() takes four principal arguments:
- the data
- cols are the names of the columns we use to fill the a new values variable (or to drop).
- the names_to column variable we wish to create from the cols provided.
- the values_to column variable we wish to create and fill with values associated with the cols provided.
R
interviews_long <- interviews_items_owned %>%
pivot_longer(cols = bicycle:car,
names_to = "items_owned",
values_to = "items_owned_logical")
View both interviews_long and
interviews_items_owned and compare their structure.
Exercise
We created some summary tables on interviews_items_owned
using count and summarise. We can create the
same tables on interviews_long, but this will require a
different process.
Make a table showing the number of respondents in each village who
owned a particular item, and include all items. The difference between
this format and the wide format is that you can now count
all the items using the items_owned variable.
R
interviews_long %>%
filter(items_owned_logical) %>%
group_by(village) %>%
count(items_owned)
OUTPUT
# A tibble: 47 × 3
# Groups: village [3]
village items_owned n
<chr> <chr> <int>
1 Chirodzo bicycle 17
2 Chirodzo computer 2
3 Chirodzo cow_cart 6
4 Chirodzo cow_plough 20
5 Chirodzo electricity 1
6 Chirodzo fridge 1
7 Chirodzo lorry 1
8 Chirodzo mobile_phone 25
9 Chirodzo motorcyle 13
10 Chirodzo no_listed_items 3
# ℹ 37 more rowsApplying what we learned to clean our data
Now we have simultaneously learned about pivot_longer()
and pivot_wider(), and fixed a problem in the way our data
is structured. In this dataset, we have another column that stores
multiple values in a single cell. Some of the cells in the
months_lack_food column contain multiple months which, as
before, are separated by semi-colons (;).
To create a data frame where each of the columns contain only one
value per cell, we can repeat the steps we applied to
items_owned and apply them to
months_lack_food. Since we will be using this data frame
for the next episode, we will call it
interviews_plotting.
R
## Plotting data ##
interviews_plotting <- interviews %>%
## pivot wider by items_owned
separate_longer_delim(items_owned, delim = ";") %>%
replace_na(list(items_owned = "no_listed_items")) %>%
## Use of grouped mutate to find number of rows
group_by(key_ID) %>%
mutate(items_owned_logical = TRUE,
number_items = if_else(items_owned == "no_listed_items", 0, n())) %>%
pivot_wider(names_from = items_owned,
values_from = items_owned_logical,
values_fill = list(items_owned_logical = FALSE)) %>%
## pivot wider by months_lack_food
separate_longer_delim(months_lack_food, delim = ";") %>%
mutate(months_lack_food_logical = TRUE,
number_months_lack_food = if_else(months_lack_food == "none", 0, n())) %>%
pivot_wider(names_from = months_lack_food,
values_from = months_lack_food_logical,
values_fill = list(months_lack_food_logical = FALSE))
Exporting data
Now that you have learned how to use
dplyr and
tidyr to wrangle your raw data, you may
want to export these new datasets to share them with your collaborators
or for archival purposes.
Similar to the read_csv() function used for reading CSV
files into R, there is a write_csv() function that
generates CSV files from data frames.
Before using write_csv(), we are going to create a new
folder, data_output, in our working directory that will
store this generated dataset. We don’t want to write generated datasets
in the same directory as our raw data. It’s good practice to keep them
separate. The data folder should only contain the raw,
unaltered data, and should be left alone to make sure we don’t delete or
modify it. In contrast, our script will generate the contents of the
data_output directory, so even if the files it contains are
deleted, we can always re-generate them.
In preparation for our next lesson on plotting, we created a version
of the dataset where each of the columns includes only one data value.
Now we can save this data frame to our data_output
directory.
R
write_csv(interviews_plotting, file = "data_output/interviews_plotting.csv")
- Use the
tidyrpackage to change the layout of data frames. - Use
pivot_wider()to go from long to wide format. - Use
pivot_longer()to go from wide to long format.
Content from Data Visualisation with ggplot2
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- What are the components of a ggplot?
- What are the main differences between R base plots, lattice, and ggplot?
- How do I create scatterplots, boxplots, and barplots?
- How can I change the aesthetics (ex. colour, transparency) of my plot?
- How can I create multiple plots at once?
Objectives
- Produce scatter plots, boxplots, and barplots using ggplot.
- Set universal plot settings.
- Describe what faceting is and apply faceting in ggplot.
- Modify the aesthetics of an existing ggplot plot (including axis labels and colour).
- Build complex and customized plots from data in a data frame.
- Recognize the differences between base R, lattice, and ggplot visualizations.
We start by loading the required package.
ggplot2 is also included in the
tidyverse package.
R
library(tidyverse)
If not still in the workspace, load the data we saved in the previous lesson.
R
interviews_plotting <- read_csv("data_output/interviews_plotting.csv")
OUTPUT
Rows: 131 Columns: 45
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (5): village, respondent_wall_type, memb_assoc, affect_conflicts, inst...
dbl (8): key_ID, no_membrs, years_liv, rooms, liv_count, no_meals, number_...
lgl (31): bicycle, television, solar_panel, table, cow_cart, radio, cow_plo...
dttm (1): interview_date
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.If you were unable to complete the previous lesson or did not save
the data, then you can create it now. Either download it using
read_csv() (Option 1) or create it with the
dplyr and tidyr code (Option 2).
Visualization Options in R
Before we start with ggplot2, it’s
helpful to know that there are several ways to create visualizations in
R. While ggplot2 is great for building
complex and highly customizable plots, there are simpler and quicker
alternatives that you might encounter or use depending on the context.
Let’s briefly explore a few of them:
R Base Plots
Base R plots are the simplest form of visualization and are great for
quick, exploratory analysis. You can create plots with very little code,
but customizing them can be cumbersome compared to
ggplot2.
Example of a simple scatterplot in base R using the
no_membrs and liv_count variables:
R
plot(interviews_plotting$no_membrs, interviews_plotting$liv_count,
main = "Base R Scatterplot",
xlab = "Number of Household Members",
ylab = "Number of Livestock Owned")
Lattice
Lattice is another plotting system in R, which allows for creating multi-panel plots easily. It’s different from ggplot2 because you define the entire plot in a single function call, and modifications after plotting are limited.
Example of a lattice plot using no_membrs and
liv_count split by village:
R
library(lattice)
R
xyplot(liv_count ~ no_membrs | village, data = interviews_plotting,
main = "Lattice Plot: Livestock Count by Household Members",
xlab = "Number of Household Members",
ylab = "Number of Livestock Owned")
Plotting with ggplot2
ggplot2 is a plotting package that
makes it simple to create complex plots from data stored in a data
frame. It provides a programmatic interface for specifying what
variables to plot, how they are displayed, and general visual
properties. Therefore, we only need minimal changes if the underlying
data change or if we decide to change from a bar plot to a scatterplot.
This helps in creating publication quality plots with minimal amounts of
adjustments and tweaking.
ggplot2 functions work best with data
in the ‘long’ format, i.e., a column for every dimension, and a row for
every observation. Well-structured data will save you lots of time when
making figures with ggplot2
ggplot graphics are built step by step by adding new elements. Adding layers in this fashion allows for extensive flexibility and customization of plots.
Each chart built with ggplot2 must include the following
Data
-
Aesthetic mapping (aes)
- Describes how variables are mapped onto graphical attributes
- Visual attribute of data including x-y axes, color, fill, shape, and alpha
-
Geometric objects (geom)
- Determines how values are rendered graphically, as bars
(
geom_bar), scatterplot (geom_point), line (geom_line), etc.
- Determines how values are rendered graphically, as bars
(
Thus, the template for graphic in ggplot2 is:
<DATA> %>%
ggplot(aes(<MAPPINGS>)) +
<GEOM_FUNCTION>()Remember from the last lesson that the pipe operator
%>% places the result of the previous line(s) into the
first argument of the function. ggplot is
a function that expects a data frame to be the first argument. This
allows for us to change from specifying the data = argument
within the ggplot function and instead pipe the data into
the function.
- use the
ggplot()function and bind the plot to a specific data frame.
R
interviews_plotting %>%
ggplot()
- define a mapping (using the aesthetic (
aes) function), by selecting the variables to be plotted and specifying how to present them in the graph, e.g. as x/y positions or characteristics such as size, shape, color, etc.
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items))
-
add ‘geoms’ – graphical representations of the data in the plot (points, lines, bars).
ggplot2offers many different geoms; we will use some common ones today, including:-
geom_point()for scatter plots, dot plots, etc. -
geom_boxplot()for, well, boxplots! -
geom_line()for trend lines, time series, etc.
-
To add a geom to the plot use the + operator. Because we
have two continuous variables, let’s use geom_point()
first:
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_point()
The + in the ggplot2
package is particularly useful because it allows you to modify existing
ggplot objects. This means you can easily set up plot
templates and conveniently explore different types of plots, so the
above plot can also be generated with code like this, similar to the
“intermediate steps” approach in the previous lesson:
R
# Assign plot to a variable
interviews_plot <- interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items))
# Draw the plot as a dot plot
interviews_plot +
geom_point()
Notes
- Anything you put in the
ggplot()function can be seen by any geom layers that you add (i.e., these are universal plot settings). This includes the x- and y-axis mapping you set up inaes(). - You can also specify mappings for a given geom independently of the
mapping defined globally in the
ggplot()function. - The
+sign used to add new layers must be placed at the end of the line containing the previous layer. If, instead, the+sign is added at the beginning of the line containing the new layer,ggplot2will not add the new layer and will return an error message.
R
## This is the correct syntax for adding layers
interviews_plot +
geom_point()
## This will not add the new layer and will return an error message
interviews_plot
+ geom_point()
Building your plots iteratively
Building plots with ggplot2 is
typically an iterative process. We start by defining the dataset we’ll
use, lay out the axes, and choose a geom:
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_point()
Then, we start modifying this plot to extract more information from
it. For instance, when inspecting the plot we notice that points only
appear at the intersection of whole numbers of no_membrs
and number_items. Also, from a rough estimate, it looks
like there are far fewer dots on the plot than there rows in our
dataframe. This should lead us to believe that there may be multiple
observations plotted on top of each other (e.g. three observations where
no_membrs is 3 and number_items is 1).
There are two main ways to alleviate overplotting issues:
- changing the transparency of the points
- jittering the location of the points
Let’s first explore option 1, changing the transparency of the
points. What we mean when we say “transparency” we mean the opacity of
point, or your ability to see through the point. We can control the
transparency of the points with the alpha argument to
geom_point. Values of alpha range from 0 to 1,
with lower values corresponding to more transparent colors (an
alpha of 1 is the default value). Specifically, an alpha of
0.1, would make a point one-tenth as opaque as a normal point. Stated
differently ten points stacked on top of each other would correspond to
a normal point.
Here, we change the alpha to 0.5, in an attempt to help
fix the overplotting. While the overplotting isn’t solved, adding
transparency begins to address this problem, as the points where there
are overlapping observations are darker (as opposed to lighter
gray):
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_point(alpha = 0.5)
That only helped a little bit with the overplotting problem, so let’s try option two. We can jitter the points on the plot, so that we can see each point in the locations where there are overlapping points. Jittering introduces a little bit of randomness into the position of our points. You can think of this process as taking the overplotted graph and giving it a tiny shake. The points will move a little bit side-to-side and up-and-down, but their position from the original plot won’t dramatically change. Note that this solution is suitable for plotting integer figures, while for numeric figures with decimals, geom_jitter() becomes inappropriate because it obscures the true value of the observation.
We can jitter our points using the geom_jitter()
function instead of the geom_point() function, as seen
below:
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_jitter()
The geom_jitter() function allows for us to specify the
amount of random motion in the jitter, using the width and
height arguments. When we don’t specify values for
width and height, geom_jitter()
defaults to 40% of the resolution of the data (the smallest change that
can be measured). Hence, if we would like less spread in our
jitter than was default, we should pick values between 0.1 and 0.4.
Experiment with the values to see how your plot changes.
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_jitter(alpha = 0.5,
width = 0.2,
height = 0.2)
For our final change, we can also add colours for all the points by
specifying a color argument inside the
geom_jitter() function:
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_jitter(alpha = 0.5,
color = "blue",
width = 0.2,
height = 0.2)
To colour each village in the plot differently, you could use a
vector as an input to the argument color.
However, because we are now mapping features of the data to a colour,
instead of setting one colour for all points, the colour of the points
now needs to be set inside a call to the
aes function. When we map a variable in
our data to the colour of the points,
ggplot2 will provide a different colour
corresponding to the different values of the variable. We will continue
to specify the value of alpha,
width, and
height outside of the
aes function because we are using the same
value for every point. ggplot2 understands both the Commonwealth English
and American English spellings for colour, i.e., you can use either
color or colour. Here is an example where we
color points by the village of the
observation:
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items)) +
geom_jitter(aes(color = village), alpha = 0.5, width = 0.2, height = 0.2)
There appears to be a positive trend between number of household members and number of items owned (from the list provided). Additionally, this trend does not appear to be different by village.
Notes
As you will learn, there are multiple ways to plot the a relationship
between variables. Another way to plot data with overlapping points is
to use the geom_count plotting function. The
geom_count() function makes the size of each point
representative of the number of data items of that type and the legend
gives point sizes associated to particular numbers of items.
R
interviews_plotting %>%
ggplot(aes(x = no_membrs, y = number_items, color = village)) +
geom_count()
Exercise
Use what you just learned to create a scatter plot of
rooms by village with the
respondent_wall_type showing in different colours. Does
this seem like a good way to display the relationship between these
variables? What other kinds of plots might you use to show this type of
data?
R
interviews_plotting %>%
ggplot(aes(x = village, y = rooms)) +
geom_jitter(aes(color = respondent_wall_type),
alpha = 0.5,
width = 0.2,
height = 0.2)
This is not a great way to show this type of data because it is difficult to distinguish between villages. What other plot types could help you visualize this relationship better?
Boxplot
We can use boxplots to visualize the distribution of rooms for each wall type:
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type, y = rooms)) +
geom_boxplot()
By adding points to a boxplot, we can have a better idea of the number of measurements and of their distribution:
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type, y = rooms)) +
geom_boxplot(alpha = 0) +
geom_jitter(alpha = 0.5,
color = "tomato",
width = 0.2,
height = 0.2)
We can see that muddaub houses and sunbrick houses tend to be smaller than burntbrick houses.
Notice how the boxplot layer is behind the jitter layer? What do you need to change in the code to put the boxplot layer in front of the jitter layer?
Exercise
Boxplots are useful summaries, but hide the shape of the distribution. For example, if the distribution is bimodal, we would not see it in a boxplot. An alternative to the boxplot is the violin plot, where the shape (of the density of points) is drawn.
- Replace the box plot with a violin plot; see
geom_violin().
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type, y = rooms)) +
geom_violin(alpha = 0) +
geom_jitter(alpha = 0.5, color = "tomato")
WARNING
Warning: Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Exercise (continued)
So far, we’ve looked at the distribution of room number within wall type. Try making a new plot to explore the distribution of another variable within wall type.
- Create a boxplot for
liv_countfor each wall type. Overlay the boxplot layer on a jitter layer to show actual measurements.
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type, y = liv_count)) +
geom_boxplot(alpha = 0) +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2)
Exercise (continued)
- Add colour to the data points on your boxplot according to whether
the respondent is a member of an irrigation association
(
memb_assoc).
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type, y = liv_count)) +
geom_boxplot(alpha = 0) +
geom_jitter(aes(color = memb_assoc), alpha = 0.5, width = 0.2, height = 0.2)
Barplots
Barplots are also useful for visualizing categorical data. By
default, geom_bar accepts a variable for x, and plots the
number of instances each value of x (in this case, wall type) appears in
the dataset.
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type)) +
geom_bar()
We can use the fill aesthetic for the
geom_bar() geom to colour bars by the portion of each count
that is from each village.
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type)) +
geom_bar(aes(fill = village))
This creates a stacked bar chart. These are generally more difficult
to read than side-by-side bars. We can separate the portions of the
stacked bar that correspond to each village and put them side-by-side by
using the position argument for geom_bar() and
setting it to “dodge”.
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type)) +
geom_bar(aes(fill = village), position = "dodge")
This is a nicer graphic, but we’re more likely to be interested in
the proportion of each housing type in each village than in the actual
count of number of houses of each type (because we might have sampled
different numbers of households in each village). To compare
proportions, we will first create a new data frame
(percent_wall_type) with a new column named “percent”
representing the percent of each house type in each village. We will
remove houses with cement walls, as there was only one in the
dataset.
R
percent_wall_type <- interviews_plotting %>%
filter(respondent_wall_type != "cement") %>%
count(village, respondent_wall_type) %>%
group_by(village) %>%
mutate(percent = (n / sum(n)) * 100) %>%
ungroup()
Now we can use this new data frame to create our plot showing the percentage of each house type in each village.
R
percent_wall_type %>%
ggplot(aes(x = village, y = percent, fill = respondent_wall_type)) +
geom_bar(stat = "identity", position = "dodge")
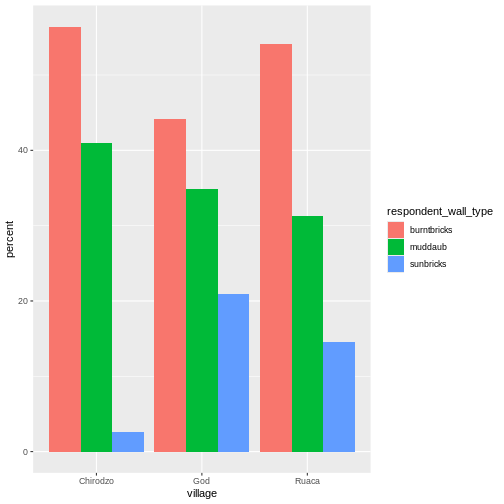
Exercise
Create a bar plot showing the proportion of respondents in each
village who are or are not part of an irrigation association
(memb_assoc). Include only respondents who answered that
question in the calculations and plot. Which village had the lowest
proportion of respondents in an irrigation association?
R
percent_memb_assoc <- interviews_plotting %>%
filter(!is.na(memb_assoc)) %>%
count(village, memb_assoc) %>%
group_by(village) %>%
mutate(percent = (n / sum(n)) * 100) %>%
ungroup()
percent_memb_assoc %>%
ggplot(aes(x = village, y = percent, fill = memb_assoc)) +
geom_bar(stat = "identity", position = "dodge")
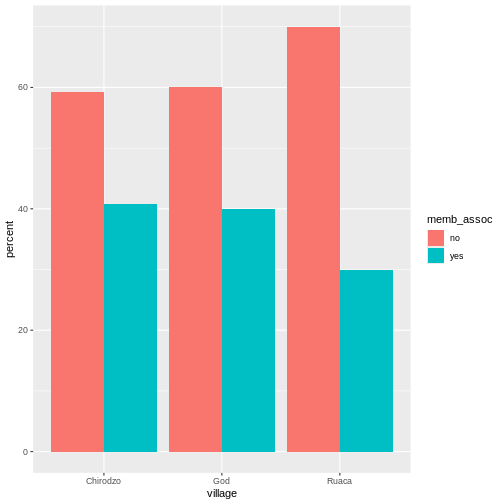
Ruaca had the lowest proportion of members in an irrigation association.
Adding Labels and Titles
By default, the axes labels on a plot are determined by the name of
the variable being plotted. However,
ggplot2 offers lots of customization
options, like specifying the axes labels, and adding a title to the plot
with relatively few lines of code. We will add more informative x-and
y-axis labels to our plot, a more explanatory label to the legend, and a
plot title.
The labs function takes the following arguments:
-
title– to produce a plot title -
subtitle– to produce a plot subtitle (smaller text placed beneath the title) -
caption– a caption for the plot -
...– any pair of name and value for aesthetics used in the plot (e.g.,x,y,fill,color,size)
R
percent_wall_type %>%
ggplot(aes(x = village, y = percent, fill = respondent_wall_type)) +
geom_bar(stat = "identity", position = "dodge") +
labs(title = "Proportion of wall type by village",
fill = "Type of Wall in Home",
x = "Village",
y = "Percent")
Faceting
Rather than creating a single plot with side-by-side bars for each village, we may want to create multiple plot, where each plot shows the data for a single village. This would be especially useful if we had a large number of villages that we had sampled, as a large number of side-by-side bars will become more difficult to read.
ggplot2 has a special technique called
faceting that allows the user to split one plot into multiple
plots based on a factor included in the dataset. We will use it to split
our barplot of housing type proportion by village so that each village
has its own panel in a multi-panel plot:
R
percent_wall_type %>%
ggplot(aes(x = respondent_wall_type, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
labs(title="Proportion of wall type by village",
x="Wall Type",
y="Percent") +
facet_wrap(~ village)
Click the “Zoom” button in your RStudio plots pane to view a larger version of this plot.
Usually plots with white background look more readable when printed.
We can set the background to white using the function
theme_bw(). Additionally, you can remove the grid:
R
percent_wall_type %>%
ggplot(aes(x = respondent_wall_type, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
labs(title="Proportion of wall type by village",
x="Wall Type",
y="Percent") +
facet_wrap(~ village) +
theme_bw() +
theme(panel.grid = element_blank())
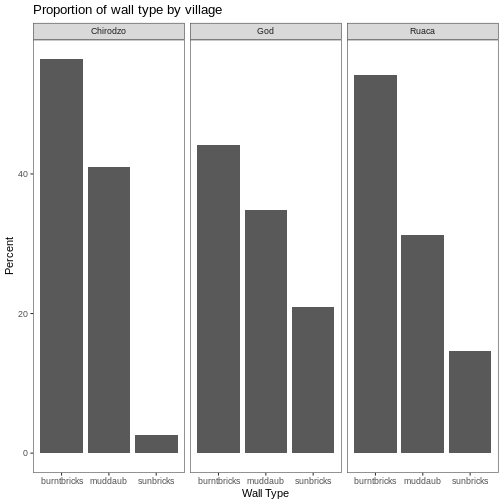
What if we wanted to see the proportion of respondents in each village who owned a particular item? We can calculate the percent of people in each village who own each item and then create a faceted series of bar plots where each plot is a particular item. First we need to calculate the percentage of people in each village who own each item:
R
percent_items <- interviews_plotting %>%
group_by(village) %>%
summarize(across(bicycle:no_listed_items, ~ sum(.x) / n() * 100)) %>%
pivot_longer(bicycle:no_listed_items, names_to = "items", values_to = "percent")
To calculate this percentage data frame, we needed to use the
across() function within a summarize()
operation. Unlike the previous example with a single wall type variable,
where each response was exactly one of the types specified, people can
(and do) own more than one item. So there are multiple columns of data
(one for each item), and the percentage calculation needs to be repeated
for each column.
Combining summarize() with across() allows
us to specify first, the columns to be summarized
(bicycle:no_listed_items) and then the calculation. Because
our calculation is a bit more complex than is available in a built-in
function, we define a new formula:
-
~indicates that we are defining a formula, -
sum(.x)gives the number of people owning that item by counting the number ofTRUEvalues (.xis shorthand for the column being operated on), - and
n()gives the current group size.
After the summarize() operation, we have a table of
percentages with each item in its own column, so a
pivot_longer() is required to transform the table into an
easier format for plotting. Using this data frame, we can now create a
multi-paneled bar plot.
R
percent_items %>%
ggplot(aes(x = village, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ items) +
theme_bw() +
theme(panel.grid = element_blank())
ggplot2 themes
In addition to theme_bw(), which changes the plot
background to white, ggplot2 comes with
several other themes which can be useful to quickly change the look of
your visualization. The complete list of themes is available at https://ggplot2.tidyverse.org/reference/ggtheme.html.
theme_minimal() and theme_light() are popular,
and theme_void() can be useful as a starting point to
create a new hand-crafted theme.
The ggthemes
package provides a wide variety of options (including an Excel 2003
theme). The ggplot2
extensions website provides a list of packages that extend the
capabilities of ggplot2, including
additional themes.
Exercise
Experiment with at least two different themes. Build the previous plot using each of those themes. Which do you like best?
Customization
Take a look at the ggplot2
cheat sheet, and think of ways you could improve the plot.
Now, let’s change names of axes to something more informative than ‘village’ and ‘percent’ and add a title to the figure:
R
percent_items %>%
ggplot(aes(x = village, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ items) +
labs(title = "Percent of respondents in each village who owned each item",
x = "Village",
y = "Percent of Respondents") +
theme_bw()
The axes have more informative names, but their readability can be improved by increasing the font size:
R
percent_items %>%
ggplot(aes(x = village, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ items) +
labs(title = "Percent of respondents in each village who owned each item",
x = "Village",
y = "Percent of Respondents") +
theme_bw() +
theme(text = element_text(size = 16))
Note that it is also possible to change the fonts of your plots. If
you are on Windows, you may have to install the extrafont
package, and follow the instructions included in the README for this
package.
After our manipulations, you may notice that the values on the x-axis are still not properly readable. Let’s change the orientation of the labels and adjust them vertically and horizontally so they don’t overlap. You can use a 90-degree angle, or experiment to find the appropriate angle for diagonally oriented labels. With a larger font, the title also runs off. We can add “\n” in the string for the title to insert a new line:
R
percent_items %>%
ggplot(aes(x = village, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ items) +
labs(title = "Percent of respondents in each village \n who owned each item",
x = "Village",
y = "Percent of Respondents") +
theme_bw() +
theme(axis.text.x = element_text(colour = "grey20", size = 12, angle = 45,
hjust = 0.5, vjust = 0.5),
axis.text.y = element_text(colour = "grey20", size = 12),
text = element_text(size = 16))
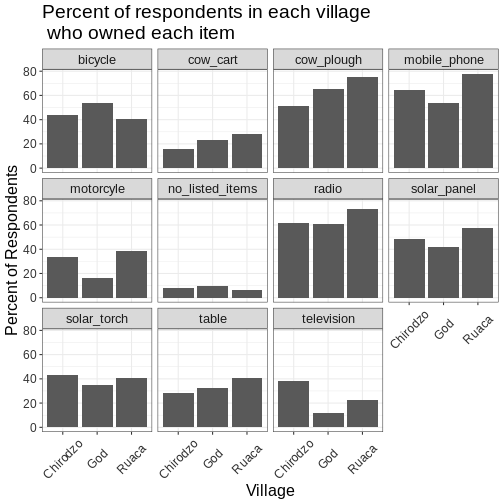
If you like the changes you created better than the default theme,
you can save them as an object to be able to easily apply them to other
plots you may create. We can also add
plot.title = element_text(hjust = 0.5) to centre the
title:
R
grey_theme <- theme(axis.text.x = element_text(colour = "grey20", size = 12,
angle = 45, hjust = 0.5,
vjust = 0.5),
axis.text.y = element_text(colour = "grey20", size = 12),
text = element_text(size = 16),
plot.title = element_text(hjust = 0.5))
percent_items %>%
ggplot(aes(x = village, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ items) +
labs(title = "Percent of respondents in each village \n who owned each item",
x = "Village",
y = "Percent of Respondents") +
grey_theme
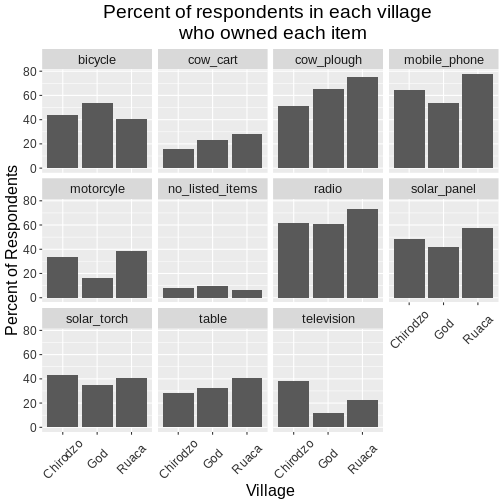
Exercise
With all of this information in hand, please take another five
minutes to either improve one of the plots generated in this exercise or
create a beautiful graph of your own. Use the RStudio ggplot2
cheat sheet for inspiration. Here are some ideas:
- See if you can make the bars white with black outline.
- Try using a different colour palette (see http://www.cookbook-r.com/Graphs/Colors_(ggplot2)/).
After creating your plot, you can save it to a file in your favourite format. The Export tab in the Plot pane in RStudio will save your plots at low resolution, which will not be accepted by many journals and will not scale well for posters.
Instead, use the ggsave() function, which allows you to
easily change the dimension and resolution of your plot by adjusting the
appropriate arguments (width, height and
dpi).
Make sure you have the fig_output/ folder in your
working directory.
R
my_plot <- percent_items %>%
ggplot(aes(x = village, y = percent)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~ items) +
labs(title = "Percent of respondents in each village \n who owned each item",
x = "Village",
y = "Percent of Respondents") +
theme_bw() +
theme(axis.text.x = element_text(color = "grey20", size = 12, angle = 45,
hjust = 0.5, vjust = 0.5),
axis.text.y = element_text(color = "grey20", size = 12),
text = element_text(size = 16),
plot.title = element_text(hjust = 0.5))
ggsave("fig_output/name_of_file.png", my_plot, width = 15, height = 10)
Note: The parameters width and height also
determine the font size in the saved plot.
-
ggplot2is a flexible and useful tool for creating plots in R. - The data set and coordinate system can be defined using the
ggplotfunction. - Additional layers, including geoms, are added using the
+operator. - Boxplots are useful for visualizing the distribution of a continuous variable.
- Barplots are useful for visualizing categorical data.
- Faceting allows you to generate multiple plots based on a categorical variable.
Content from Getting started with R Markdown (Optional)
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- What is R Markdown?
- How can I integrate my R code with text and plots?
- How can I convert .Rmd files to .html?
Objectives
- Create a .Rmd document containing R code, text, and plots
- Create a YAML header to control output
- Understand basic syntax of (R)Markdown
- Customise code chunks to control formatting
- Use code chunks and in-line code to create dynamic, reproducible documents
R Markdown
R Markdown is a flexible type of document that allows you to seamlessly combine executable R code, and its output, with text in a single document. These documents can be readily converted to multiple static and dynamic output formats, including PDF (.pdf), Word (.docx), and HTML (.html).
The benefit of a well-prepared R Markdown document is full reproducibility. This also means that, if you notice a data transcription error, or you are able to add more data to your analysis, you will be able to recompile the report without making any changes in the actual document.
The rmarkdown package comes pre-installed with RStudio, so no action is necessary.

Creating an R Markdown file
To create a new R Markdown document in RStudio, click File -> New File -> R Markdown:
Then click on ‘Create Empty Document’. Normally you could enter the title of your document, your name (Author), and select the type of output, but we will be learning how to start from a blank document.
Basic components of R Markdown
To control the output, a YAML (YAML Ain’t Markup Language) header is needed:
---
title: "My Awesome Report"
author: "Emmet Brickowski"
date: ""
output: html_document
---The header is defined by the three hyphens at the beginning
(---) and the three hyphens at the end
(---).
In the YAML, the only required field is the output:,
which specifies the type of output you want. This can be an
html_document, a pdf_document, or a
word_document. We will start with an HTML doument and
discuss the other options later.
The rest of the fields can be deleted, if you don’t need them. After
the header, to begin the body of the document, you start typing after
the end of the YAML header (i.e. after the second ---).
Markdown syntax
Markdown is a popular markup language that allows you to add
formatting elements to text, such as bold,
italics, and code. The formatting will not be
immediately visible in a markdown (.md) document, like you would see in
a Word document. Rather, you add Markdown syntax to the text, which can
then be converted to various other files that can translate the Markdown
syntax. Markdown is useful because it is lightweight, flexible, and
platform independent.
Some platforms provide a real time preview of the formatting, like RStudio’s visual markdown editor (available from version 1.4).
First, let’s create a heading! A # in front of text
indicates to Markdown that this text is a heading. Adding more
#s make the heading smaller, i.e. one # is a
first level heading, two ##s is a second level heading,
etc. upto the 6th level heading.
# Title
## Section
### Sub-section
#### Sub-sub section
##### Sub-sub-sub section
###### Sub-sub-sub-sub section(only use a level if the one above is also in use)
Since we have already defined our title in the YAML header, we will use a section heading to create an Introduction section.
## IntroductionYou can make things bold by surrounding the word
with double asterisks, **bold**, or double underscores,
__bold__; and italicize using single asterisks,
*italics*, or single underscores,
_italics_.
You can also combine bold and italics to
write something really important with
triple-asterisks, ***really***, or underscores,
___really___; and, if you’re feeling bold (pun intended),
you can also use a combination of asterisks and underscores,
**_really_**, **_really_**.
To create code-type font, surround the word with
backticks, `code-type`.
Now that we’ve learned a couple of things, it might be useful to implement them:
## Introduction
This report uses the **tidyverse** package along with the *SAFI* dataset,
which has columns that include:Then we can create a list for the variables using -,
+, or * keys.
## Introduction
This report uses the **tidyverse** package along with the *SAFI* dataset,
which has columns that include:
- village
- interview_date
- no_members
- years_liv
- respondent_wall_type
- roomsYou can also create an ordered list using numbers:
1. village
2. interview_date
3. no_members
4. years_liv
5. respondent_wall_type
6. roomsAnd nested items by tab-indenting:
- village
+ Name of village
- interview_date
+ Date of interview
- no_members
+ How many family members lived in a house
- years_liv
+ How many years respondent has lived in village or neighbouring village
- respondent_wall_type
+ Type of wall of house
- rooms
+ Number of rooms in houseFor more Markdown syntax see the following reference guide.
Now we can render the document into HTML by clicking the Knit button in the top of the Source pane (top left), or use the keyboard shortcut Ctrl+Shift+K on Windows and Linux, and Cmd+Shift+K on Mac. If you haven’t saved the document yet, you will be prompted to do so when you Knit for the first time.
Writing an R Markdown report
Now we will add some R code from our previous data wrangling and visualisation, which means we need to make sure tidyverse is loaded. It is not enough to load tidyverse from the console, we will need to load it within our R Markdown document. The same applies to our data. To load these, we will need to create a ‘code chunk’ at the top of our document (below the YAML header).
A code chunk can be inserted by clicking Code > Insert Chunk, or by using the keyboard shortcuts Ctrl+Alt+I on Windows and Linux, and Cmd+Option+I on Mac.
The syntax of a code chunk is:
An R Markdown document knows that this text is not part of the report
from the ``` that begins and ends the chunk. It also knows
that the code inside of the chunk is R code from the r
inside of the curly braces ({}). After the r
you can add a name for the code chunk . Naming a chunk is optional, but
recommended. Each chunk name must be unique, and only contain
alphanumeric characters and -.
To load tidyverse and our
SAFI_clean.csv file, we will insert a chunk and call it
‘setup’. Since we don’t want this code or the output to show in our
knitted HTML document, we add an include = FALSE option
after the code chunk name ({r setup, include = FALSE}).
MARKDOWN
```{r setup, include = FALSE}
library(tidyverse)
library(here)
interviews <- read_csv(here("data/SAFI_clean.csv"), na = "NULL")
```Important Note!
The file paths you give in a .Rmd document, e.g. to load a .csv file, are relative to the .Rmd document, not the project root.
As suggested in the Starting with Data episode, we highly recommend
the use of the here() function to keep the file paths
consistent within your project.
Insert table
Next, we will re-create a table from the Data Wrangling episode which
shows the average household size grouped by village and
memb_assoc. We can do this by creating a new code chunk and
calling it ‘interview-tbl’. Or, you can come up with something more
creative (just remember to stick to the naming rules).
It isn’t necessary to Knit your document every time you want to see the output. Instead you can run the code chunk with the green triangle in the top right corner of the the chunk, or with the keyboard shortcuts: Ctrl+Alt+C on Windows and Linux, or Cmd+Option+C on Mac.
To make sure the table is formatted nicely in our output document, we
will need to use the kable() function from the
knitr package. The kable() function takes
the output of your R code and knits it into a nice looking HTML table.
You can also specify different aspects of the table, e.g. the column
names, a caption, etc.
Run the code chunk to make sure you get the desired output.
R
interviews %>%
filter(!is.na(memb_assoc)) %>%
group_by(village, memb_assoc) %>%
summarize(mean_no_membrs = mean(no_membrs)) %>%
knitr::kable(caption = "We can also add a caption.",
col.names = c("Village", "Member Association",
"Mean Number of Members"))
| Village | Member Association | Mean Number of Members |
|---|---|---|
| Chirodzo | no | 8.062500 |
| Chirodzo | yes | 7.818182 |
| God | no | 7.133333 |
| God | yes | 8.000000 |
| Ruaca | no | 7.178571 |
| Ruaca | yes | 9.500000 |
Many different R packages can be used to generate tables. Some of the more commonly used options are listed in the table below.
| Name | Creator(s) | Description |
|---|---|---|
| condformat | Oller Moreno (2022) | Apply and visualize conditional formatting to data frames in R. It renders a data frame with cells formatted according to criteria defined by rules, using a tidy evaluation syntax. |
| DT | Xie et al. (2023) | Data objects in R can be rendered as HTML tables using the JavaScript library ‘DataTables’ (typically via R Markdown or Shiny). The ‘DataTables’ library has been included in this R package. |
| formattable | Ren and Russell (2021) | Provides functions to create formattable vectors and data frames. ‘Formattable’ vectors are printed with text formatting, and formattable data frames are printed with multiple types of formatting in HTML to improve the readability of data presented in tabular form rendered on web pages. |
| flextable | Gohel and Skintzos (2023) | Use a grammar for creating and customizing pretty tables. The following formats are supported: ‘HTML’, ‘PDF’, ‘RTF’, ‘Microsoft Word’, ‘Microsoft PowerPoint’ and R ‘Grid Graphics’. ‘R Markdown’, ‘Quarto’, and the package ‘officer’ can be used to produce the result files. |
| gt | Iannone et al. (2022) | Build display tables from tabular data with an easy-to-use set of functions. With its progressive approach, we can construct display tables with cohesive table parts. Table values can be formatted using any of the included formatting functions. |
| huxtable | Hugh-Jones (2022) | Creates styled tables for data presentation. Export to HTML, LaTeX, RTF, ‘Word’, ‘Excel’, and ‘PowerPoint’. Simple, modern interface to manipulate borders, size, position, captions, colours, text styles and number formatting. |
| pander | Daróczi and Tsegelskyi (2022) | Contains some functions catching all messages, ‘stdout’ and other useful information while evaluating R code and other helpers to return user specified text elements (e.g., header, paragraph, table, image, lists etc.) in ‘pandoc’ markdown or several types of R objects similarly automatically transformed to markdown format. |
| pixiedust | Nutter and Kretch (2021) | ‘pixiedust’ provides tidy data frames with a programming interface intended to be similar to ’ggplot2’s system of layers with fine-tuned control over each cell of the table. |
| reactable | Lin et al. (2023) | Interactive data tables for R, based on the ‘React Table’ JavaScript library. Provides an HTML widget that can be used in ‘R Markdown’ or ‘Quarto’ documents, ‘Shiny’ applications, or viewed from an R console. |
| rhandsontable | Owen et al. (2021) | An R interface to the ‘Handsontable’ JavaScript library, which is a minimalist Excel-like data grid editor. |
| stargazer | Hlavac (2022) | Produces LaTeX code, HTML/CSS code and ASCII text for well-formatted tables that hold regression analysis results from several models side-by-side, as well as summary statistics. |
| tables | Murdoch (2022) | Computes and displays complex tables of summary statistics. Output may be in LaTeX, HTML, plain text, or an R matrix for further processing. |
| tangram | Garbett et al. (2023) | Provides an extensible formula system to quickly and easily create production quality tables. The processing steps are a formula parser, statistical content generation from data defined by a formula, and rendering into a table. |
| xtable | Dahl et al. (2019) | Coerce data to LaTeX and HTML tables. |
| ztable | Moon (2021) | Makes zebra-striped tables (tables with alternating row colors) in LaTeX and HTML formats easily from a data.frame, matrix, lm, aov, anova, glm, coxph, nls, fitdistr, mytable and cbind.mytable objects. |
Customising chunk output
We mentioned using include = FALSE in a code chunk to
prevent the code and output from printing in the knitted document. There
are additional options available to customise how the code-chunks are
presented in the output document. The options are entered in the code
chunk after chunk-name and separated by commas,
e.g. {r chunk-name, eval = FALSE, echo = TRUE}.
| Option | Options | Output |
|---|---|---|
eval |
TRUE or FALSE
|
Whether or not the code within the code chunk should be run. |
echo |
TRUE or FALSE
|
Choose if you want to show your code chunk in the output document.
echo = TRUE will show the code chunk. |
include |
TRUE or FALSE
|
Choose if the output of a code chunk should be included in the
document. FALSE means that your code will run, but will not
show up in the document. |
warning |
TRUE or FALSE
|
Whether or not you want your output document to display potential warning messages produced by your code. |
message |
TRUE or FALSE
|
Whether or not you want your output document to display potential messages produced by your code. |
fig.align |
default, left, right,
center
|
Where the figure from your R code chunk should be output on the page |
Tip
- The default settings for the above chunk options are all
TRUE. - The default settings can be modified per chunk, or with
knitr::opts_chunk$set(), - Entering
knitr::opts_chunk$set(echo = FALSE)will change the default of value ofechotoFALSEfor every code chunk in the document.
Exercise
Play around with the different options in the chunk with the code for the table, and re-Knit to see what each option does to the output.
What happens if you use eval = FALSE and
echo = FALSE? What is the difference between this and
include = FALSE?
Create a chunk with {r eval = FALSE, echo = FALSE}, then
create another chunk with {r include = FALSE} to compare.
eval = FALSE and echo = FALSE will neither run
the code in the chunk, nor show the code in the knitted document. The
code chunk essentially doesn’t exist in the knitted document as it was
never run. Whereas include = FALSE will run the code and
store the output for later use.
In-line R code
Now we will use some in-line R code to present some descriptive
statistics. To use in-line R-code, we use the same backticks that we
used in the Markdown section, with an r to specify that we
are generating R-code. The difference between in-line code and a code
chunk is the number of backticks. In-line R code uses one backtick
(`r`), whereas code chunks use three backticks
(```r```).
For example, today’s date is `r Sys.Date()`, will be
rendered as: today’s date is 2026-01-27.
The code will display today’s date in the output document (well,
technically the date the document was last knitted).
The best way to use in-line R code, is to minimise the amount of code you need to produce the in-line output by preparing the output in code chunks. Let’s say we’re interested in presenting the average household size in a village.
R
# create a summary data frame with the mean household size by village
mean_household <- interviews %>%
group_by(village) %>%
summarize(mean_no_membrs = mean(no_membrs))
# and select the village we want to use
mean_chirodzo <- mean_household %>%
filter(village == "Chirodzo")
Now we can make an informative statement on the means of each village, and include the mean values as in-line R-code. For example:
The average household size in the village of Chirodzo is
`r round(mean_chirodzo$mean_no_membrs, 2)`
becomes…
The average household size in the village of Chirodzo is 7.08.
Because we are using in-line R code instead of the actual values, we have created a dynamic document that will automatically update if we make changes to the dataset and/or code chunks.
Plots
Finally, we will also include a plot, so our document is a little
more colourful and a little less boring. We will use the
interview_plotting data from the previous episode.
If you were unable to complete the previous lesson or did not save the data, then you can create it in a new code chunk.
R
## Not run, but can be used to load in data from previous lesson!
interviews_plotting <- interviews %>%
## pivot wider by items_owned
separate_rows(items_owned, sep = ";") %>%
## if there were no items listed, changing NA to no_listed_items
replace_na(list(items_owned = "no_listed_items")) %>%
mutate(items_owned_logical = TRUE) %>%
pivot_wider(names_from = items_owned,
values_from = items_owned_logical,
values_fill = list(items_owned_logical = FALSE)) %>%
## pivot wider by months_lack_food
separate_rows(months_lack_food, sep = ";") %>%
mutate(months_lack_food_logical = TRUE) %>%
pivot_wider(names_from = months_lack_food,
values_from = months_lack_food_logical,
values_fill = list(months_lack_food_logical = FALSE)) %>%
## add some summary columns
mutate(number_months_lack_food = rowSums(select(., Jan:May))) %>%
mutate(number_items = rowSums(select(., bicycle:car)))
Exercise
Create a new code chunk for the plot, and copy the code from any of the plots we created in the previous episode to produce a plot in the chunk. I recommend one of the colourful plots.
If you are feeling adventurous, you can also create a new plot with
the interviews_plotting data frame.
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type)) +
geom_bar(aes(fill = village))
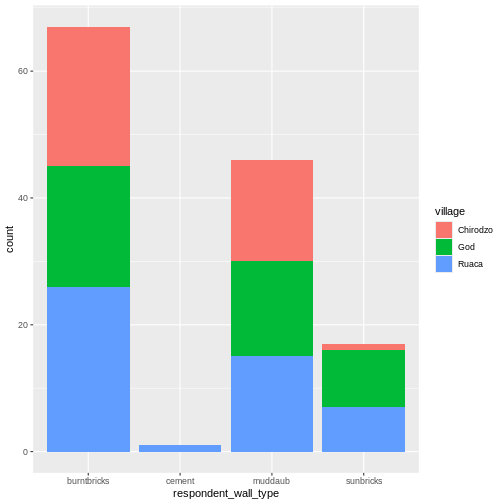
We can also create a caption with the chunk option
fig.cap.
MARKDOWN
```{r chunk-name, fig.cap = "I made this plot while attending an
awesome Data Carpentries workshop where I learned a ton of cool stuff!"}
Code for plot
```…or, ideally, something more informative.
R
interviews_plotting %>%
ggplot(aes(x = respondent_wall_type)) +
geom_bar(aes(fill = village), position = "dodge") +
labs(x = "Type of Wall in Home", y = "Count", fill = "Village Name") +
scale_fill_viridis_d() # add colour deficient friendly palette
Other output options
You can convert R Markdown to a PDF or a Word document (among
others). Click the little triangle next to the Knit
button to get a drop-down menu. Or you could put
pdf_document or word_document in the initial
header of the file.
---
title: "My Awesome Report"
author: "Emmet Brickowski"
date: ""
output: word_document
---Note: Creating PDF documents
Creating .pdf documents may require installation of some extra
software. The R package tinytex provides some tools to help
make this process easier for R users. With tinytex
installed, run tinytex::install_tinytex() to install the
required software (you’ll only need to do this once) and then when you
Knit to pdf tinytex will automatically
detect and install any additional LaTeX packages that are needed to
produce the pdf document. Visit the tinytex website for more
information.
Note: Inserting citations into an R Markdown file
It is possible to insert citations into an R Markdown file using the
editor toolbar. The editor toolbar includes commonly seen formatting
buttons generally seen in text editors (e.g., bold and italic buttons).
The toolbar is accessible by using the settings dropdown menu (next to
the ‘Knit’ dropdown menu) to select ‘Use Visual Editor’, also accessible
through the shortcut ‘Crtl+Shift+F4’. From here, clicking ‘Insert’
allows ‘Citation’ to be selected (shortcut: ‘Crtl+Shift+F8’). For
example, searching ‘10.1007/978-3-319-24277-4’ in ‘From DOI’ and
inserting will provide the citation for ggplot2 [@wickham2016]. This will also save the
citation(s) in ‘references.bib’ in the current working directory. Visit
the R
Studio website for more information. Tip: obtaining citation
information from relevant packages can be done by using
citation("package").
Resources
- Knitr in a knutshell tutorial
- Dynamic Documents with R and knitr (book)
- R Markdown documentation
- R Markdown cheat sheet
- Getting started with R Markdown
- Markdown tutorial
- R Markdown: The Definitive Guide (book by Rstudio team)
- Reproducible Reporting
- Introducing Bookdown
- R Markdown is a useful language for creating reproducible documents combining text and executable R-code.
- Specify chunk options to control formatting of the output document
Content from Processing JSON data (Optional)
Last updated on 2026-01-27 | Edit this page
Overview
Questions
- What is JSON format?
- How can I convert JSON to an R dataframe?
- How can I convert an array of JSON record into a table?
Objectives
- Describe the JSON data format
- Understand where JSON is typically used
- Appreciate some advantages of using JSON over tabular data
- Appreciate some disadvantages of processing JSON documents
- Use the jsonLite package to read a JSON file
- Display formatted JSON as dataframe
- Select and display nested dataframe fields from a JSON document
- Write tabular data from selected elements from a JSON document to a csv file
The JSON data format
The JSON data format was designed as a way of allowing different machines or processes within machines to communicate with each other by sending messages constructed in a well defined format. JSON is now the preferred data format used by APIs (Application Programming Interfaces).
The JSON format although somewhat verbose is not only Human readable but it can also be mapped very easily to an R dataframe.
We are going to read a file of data formatted as JSON, convert it into a dataframe in R then selectively create a csv file from the extracted data.
The JSON file we are going to use is the SAFI.json file. This is the output file from an electronic survey system called ODK. The JSON represents the answers to a series of survey questions. The questions themselves have been replaced with unique Keys, the values are the answers.
Because detailed surveys are by nature nested structures making it possible to record different levels of detail or selectively ask a set of specific questions based on the answer given to a previous question, the structure of the answers for the survey can not only be complex and convoluted, it could easily be different from one survey respondent’s set of answers to another.
Advantages of JSON
- Very popular data format for APIs (e.g. results from an Internet search)
- Human readable
- Each record (or document as they are called) is self contained. The equivalent of the column name and column values are in every record.
- Documents do not all have to have the same structure within the same file
- Document structures can be complex and nested
Use the JSON package to read a JSON file
R
library(jsonlite)
As with reading in a CSV, you have a couple of options for how to access the JSON file.
You can read the JSON file directly into R with
read_json() or the comparable fromJSON()
function, though this does not download the file.
R
json_data <- read_json(
"https://raw.githubusercontent.com/datacarpentry/r-socialsci/main/episodes/data/SAFI.json"
)
To download the file you can copy and paste the contents of the file
on GitHub,
creating a SAFI.json file in your data
directory, or you can download the file with R.
R
download.file(
"https://raw.githubusercontent.com/datacarpentry/r-socialsci/main/episodes/data/SAFI.json",
"data/SAFI.json", mode = "wb")
Once you have the data downloaded, you can read it into R with
read_json():
R
json_data <- read_json("data/SAFI.json")
We can see that a new object called json_data has appeared in our
Environment. It is described as a Large list (131 elements). In this
current form, our data is messy. You can have a glimpse of it with the
head() or view() functions. It will look not
much more structured than if you were to open the JSON file with a text
editor.
This is because, by default, the read_json() function’s
parameter simplifyVector, which specifies whether or not to
simplify vectors is set to FALSE. This means that the default setting
does not simplify nested lists into vectors and data frames. However, we
can set this to TRUE, and our data will be read directly as a
dataframe:
R
json_data <- read_json("data/SAFI.json", simplifyVector = TRUE)
Now we can see we have this json data in a dataframe format. For
consistency with the rest of the lesson, let’s coerce it to be a tibble
and use glimpse to take a peek inside (these functions were
loaded by library(tidyverse)):
R
json_data <- json_data %>% as_tibble()
glimpse(json_data)
OUTPUT
Rows: 131
Columns: 74
$ C06_rooms <int> 1, 1, 1, 1, 1, 1, 1, 3, 1, 5, 1, 3, 1, …
$ B19_grand_liv <chr> "no", "yes", "no", "no", "yes", "no", "…
$ A08_ward <chr> "ward2", "ward2", "ward2", "ward2", "wa…
$ E01_water_use <chr> "no", "yes", "no", "no", "no", "no", "y…
$ B18_sp_parents_liv <chr> "yes", "yes", "no", "no", "no", "no", "…
$ B16_years_liv <int> 4, 9, 15, 6, 40, 3, 38, 70, 6, 23, 20, …
$ E_yes_group_count <chr> NA, "3", NA, NA, NA, NA, "4", "2", "3",…
$ F_liv <list> [<data.frame[1 x 2]>], [<data.frame[3 …
$ `_note2` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ instanceID <chr> "uuid:ec241f2c-0609-46ed-b5e8-fe575f6ce…
$ B20_sp_grand_liv <chr> "yes", "yes", "no", "no", "no", "no", "…
$ F10_liv_owned_other <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ `_note1` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ F12_poultry <chr> "yes", "yes", "yes", "yes", "yes", "no"…
$ D_plots_count <chr> "2", "3", "1", "3", "2", "1", "4", "2",…
$ C02_respondent_wall_type_other <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ C02_respondent_wall_type <chr> "muddaub", "muddaub", "burntbricks", "b…
$ C05_buildings_in_compound <int> 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 2, 2, 1, …
$ `_remitters` <list> [<data.frame[0 x 0]>], [<data.frame[0 …
$ E18_months_no_water <list> <NULL>, <"Aug", "Sept">, <NULL>, <NULL…
$ F07_use_income <chr> NA, "Alimentação e pagamento de educa…
$ G01_no_meals <int> 2, 2, 2, 2, 2, 2, 3, 2, 3, 3, 2, 3, 2, …
$ E17_no_enough_water <chr> NA, "yes", NA, NA, NA, NA, "yes", "yes"…
$ F04_need_money <chr> NA, "no", NA, NA, NA, NA, "no", "no", "…
$ A05_end <chr> "2017-04-02T17:29:08.000Z", "2017-04-02…
$ C04_window_type <chr> "no", "no", "yes", "no", "no", "no", "n…
$ E21_other_meth <chr> NA, "no", NA, NA, NA, NA, "no", "no", "…
$ D_no_plots <int> 2, 3, 1, 3, 2, 1, 4, 2, 3, 2, 2, 2, 4, …
$ F05_money_source <list> <NULL>, <NULL>, <NULL>, <NULL>, <NULL>…
$ A07_district <chr> "district1", "district1", "district1", …
$ C03_respondent_floor_type <chr> "earth", "earth", "cement", "earth", "e…
$ E_yes_group <list> [<data.frame[0 x 0]>], [<data.frame[3 …
$ A01_interview_date <chr> "2016-11-17", "2016-11-17", "2016-11-17…
$ B11_remittance_money <chr> "no", "no", "no", "no", "no", "no", "no…
$ A04_start <chr> "2017-03-23T09:49:57.000Z", "2017-04-02…
$ D_plots <list> [<data.frame[2 x 8]>], [<data.frame[3 …
$ F_items <list> [<data.frame[3 x 3]>], [<data.frame[2 …
$ F_liv_count <chr> "1", "3", "1", "2", "4", "1", "1", "2",…
$ F10_liv_owned <list> "poultry", <"oxen", "cows", "goats">, …
$ B_no_membrs <int> 3, 7, 10, 7, 7, 3, 6, 12, 8, 12, 6, 7, …
$ F13_du_look_aftr_cows <chr> "no", "no", "no", "no", "no", "no", "no…
$ E26_affect_conflicts <chr> NA, "once", NA, NA, NA, NA, "never", "n…
$ F14_items_owned <list> <"bicycle", "television", "solar_panel…
$ F06_crops_contr <chr> NA, "more_half", NA, NA, NA, NA, "more_…
$ B17_parents_liv <chr> "no", "yes", "no", "no", "yes", "no", "…
$ G02_months_lack_food <list> "Jan", <"Jan", "Sept", "Oct", "Nov", "…
$ A11_years_farm <dbl> 11, 2, 40, 6, 18, 3, 20, 16, 16, 22, 6,…
$ F09_du_labour <chr> "no", "no", "yes", "yes", "no", "yes", …
$ E_no_group_count <chr> "2", NA, "1", "3", "2", "1", NA, NA, NA…
$ E22_res_change <list> <NULL>, <NULL>, <NULL>, <NULL>, <NULL>…
$ E24_resp_assoc <chr> NA, "no", NA, NA, NA, NA, NA, "yes", NA…
$ A03_quest_no <chr> "01", "01", "03", "04", "05", "6", "7",…
$ `_members` <list> [<data.frame[3 x 12]>], [<data.frame[7…
$ A06_province <chr> "province1", "province1", "province1", …
$ `gps:Accuracy` <dbl> 14, 19, 13, 5, 10, 12, 11, 9, 11, 14, 1…
$ E20_exper_other <chr> NA, "yes", NA, NA, NA, NA, "yes", "yes"…
$ A09_village <chr> "village2", "village2", "village2", "vi…
$ C01_respondent_roof_type <chr> "grass", "grass", "mabatisloping", "mab…
$ `gps:Altitude` <dbl> 698, 690, 674, 679, 689, 692, 709, 700,…
$ `gps:Longitude` <dbl> 33.48346, 33.48342, 33.48345, 33.48342,…
$ E23_memb_assoc <chr> NA, "yes", NA, NA, NA, NA, "no", "yes",…
$ E19_period_use <dbl> NA, 2, NA, NA, NA, NA, 10, 10, 6, 22, N…
$ E25_fees_water <chr> NA, "no", NA, NA, NA, NA, "no", "no", "…
$ C07_other_buildings <chr> "no", "no", "no", "no", "no", "no", "ye…
$ observation <chr> "None", "Estes primeiros inquéritos na…
$ `_note` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ A12_agr_assoc <chr> "no", "yes", "no", "no", "no", "no", "n…
$ G03_no_food_mitigation <list> <"na", "rely_less_food", "reduce_meals…
$ F05_money_source_other <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ `gps:Latitude` <dbl> -19.11226, -19.11248, -19.11211, -19.11…
$ E_no_group <list> [<data.frame[2 x 6]>], [<data.frame[0 …
$ F14_items_owned_other <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ F08_emply_lab <chr> "no", "yes", "no", "no", "no", "no", "n…
$ `_members_count` <chr> "3", "7", "10", "7", "7", "3", "6", "12…Looking good, but you might notice that actually we have a variable, F_liv that is a list of dataframes! It is very important to know what you are expecting from your data to be able to look for things like this. For example, if you are getting your JSON from an API, have a look at the API documentation, so you know what to look for.
Often when we have a very large number of columns, it can become
difficult to determine all the variables which may require some special
attention, like lists. Fortunately, we can use special verbs like
where to quickly select all the list columns.
R
json_data %>%
select(where(is.list)) %>%
glimpse()
OUTPUT
Rows: 131
Columns: 14
$ F_liv <list> [<data.frame[1 x 2]>], [<data.frame[3 x 2]>], …
$ `_remitters` <list> [<data.frame[0 x 0]>], [<data.frame[0 x 0]>], …
$ E18_months_no_water <list> <NULL>, <"Aug", "Sept">, <NULL>, <NULL>, <NULL…
$ F05_money_source <list> <NULL>, <NULL>, <NULL>, <NULL>, <NULL>, <NULL>…
$ E_yes_group <list> [<data.frame[0 x 0]>], [<data.frame[3 x 14]>],…
$ D_plots <list> [<data.frame[2 x 8]>], [<data.frame[3 x 8]>], …
$ F_items <list> [<data.frame[3 x 3]>], [<data.frame[2 x 3]>], …
$ F10_liv_owned <list> "poultry", <"oxen", "cows", "goats">, "none", …
$ F14_items_owned <list> <"bicycle", "television", "solar_panel", "tabl…
$ G02_months_lack_food <list> "Jan", <"Jan", "Sept", "Oct", "Nov", "Dec">, <…
$ E22_res_change <list> <NULL>, <NULL>, <NULL>, <NULL>, <NULL>, <NULL>…
$ `_members` <list> [<data.frame[3 x 12]>], [<data.frame[7 x 12]>]…
$ G03_no_food_mitigation <list> <"na", "rely_less_food", "reduce_meals", "day_…
$ E_no_group <list> [<data.frame[2 x 6]>], [<data.frame[0 x 0]>], …So what can we do about F_liv, the column of dataframes?
Well first things first, we can access each one. For example to access
the dataframe in the first row, we can use the bracket ([)
subsetting. Here we use single bracket, but you could also use double
bracket ([[). The [[ form allows only a single
element to be selected using integer or character indices, whereas
[ allows indexing by vectors.
R
json_data$F_liv[1]
OUTPUT
[[1]]
F11_no_owned F_curr_liv
1 1 poultryWe can also choose to view the nested dataframes at all the rows of our main dataframe where a particular condition is met (for example where the value for the variable C06_rooms is equal to 4):
R
json_data$F_liv[which(json_data$C06_rooms == 4)]
OUTPUT
[[1]]
F11_no_owned F_curr_liv
1 3 oxen
2 2 cows
3 5 goats
[[2]]
F11_no_owned F_curr_liv
1 4 oxen
2 5 cows
3 3 goats
[[3]]
data frame with 0 columns and 0 rows
[[4]]
F11_no_owned F_curr_liv
1 4 oxen
2 4 cows
3 4 goats
4 1 sheep
[[5]]
F11_no_owned F_curr_liv
1 2 cowsWrite the JSON file to csv
If we try to write our json_data dataframe to a csv as we would
usually in a regular dataframe, we won’t get the desired result. Using
the write_csv function from the readr
package won’t give you an error for list columns, but you’ll only see
missing (i.e. NA) values in these columns. Let’s try it out
to confirm:
R
write_csv(json_data, "json_data_with_list_columns.csv")
read_csv("json_data_with_list_columns.csv")
To write out as a csv while maintaining the data within the list
columns, we will need to “flatten” these columns. One way to do this is
to convert these list columns into character types. (However, we don’t
want to change the data types for any of the other columns). Here’s one
way to do this using tidyverse. This command only applies the
as.character command to those columns ‘where’
is.list is TRUE.
R
flattened_json_data <- json_data %>%
mutate(across(where(is.list), as.character))
flattened_json_data
OUTPUT
# A tibble: 131 × 74
C06_rooms B19_grand_liv A08_ward E01_water_use B18_sp_parents_liv
<int> <chr> <chr> <chr> <chr>
1 1 no ward2 no yes
2 1 yes ward2 yes yes
3 1 no ward2 no no
4 1 no ward2 no no
5 1 yes ward2 no no
6 1 no ward2 no no
7 1 yes ward2 yes no
8 3 yes ward1 yes yes
9 1 yes ward2 yes no
10 5 no ward2 yes no
# ℹ 121 more rows
# ℹ 69 more variables: B16_years_liv <int>, E_yes_group_count <chr>,
# F_liv <chr>, `_note2` <lgl>, instanceID <chr>, B20_sp_grand_liv <chr>,
# F10_liv_owned_other <lgl>, `_note1` <lgl>, F12_poultry <chr>,
# D_plots_count <chr>, C02_respondent_wall_type_other <lgl>,
# C02_respondent_wall_type <chr>, C05_buildings_in_compound <int>,
# `_remitters` <chr>, E18_months_no_water <chr>, F07_use_income <chr>, …Now you can write this to a csv file:
R
write_csv(flattened_json_data, "data_output/json_data_with_flattened_list_columns.csv")
Note: this means that when you read this csv back into R, the column of the nested dataframes will now be read in as a character vector. Converting it back to list to extract elements might be complicated, so it is probably better to keep storing these data in a JSON format if you will have to do this.
You can also write out the individual nested dataframes to a csv. For example:
R
write_csv(json_data$F_liv[[1]], "data_output/F_liv_row1.csv")
- JSON is a popular data format for transferring data used by a great many Web based APIs
- The complex structure of a JSON document means that it cannot easily be ‘flattened’ into tabular data
- We can use R code to extract values of interest and place them in a csv file
